02Q PDF
Diunggah oleh
FCiênciasDeskripsi Asli:
Judul Asli
Hak Cipta
Format Tersedia
Bagikan dokumen Ini
Apakah menurut Anda dokumen ini bermanfaat?
Apakah konten ini tidak pantas?
Laporkan Dokumen IniHak Cipta:
Format Tersedia
02Q PDF
Diunggah oleh
FCiênciasHak Cipta:
Format Tersedia
Question 2
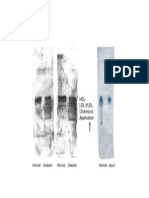
You are studying a virus called DK15, which is known to cause tumors in mice. The genome of DK15 is a 5.3 kb, double-stranded, circular DNA molecule. You are interested in learning more about DK15, so you decide to map its DNA using restriction enzymes. As a first step in your analysis, you digest DK15 DNA with three enzymes: SspI, XhoI, and SmaI. You run the digested DNA on an agarose gel, along with an uncut sample of the DK15 DNA as a control. You obtain the following pattern of bands**:
**You may assume that all the digests worked (i.e. all enzymes were "active") a) Fill in the blanks with the identities of the uncut DNA bands labeled A, B, and C: A___________________ B___________________ C___________________ b) How many times does XhoI cut the DK15 DNA? Justify your answer in one sentence. c) How many times does SspI cut the DK15 DNA? How do you know?
You are a bit puzzled by the SmaI result, and show your gel to Kate to get her input. Kate concludes that the DK15 DNA contains two SmaI siteseven though you observed three bands on your gel. d) Assuming that Kate is correct, propose a simple explanation for your observation of three bands. Explain your answer by stating the probable identity of each band you observed, and how you deduced this. To determine the restriction map of DK15 DNA, you perform the following complete digests with XhoI, SmaI, and a third enzyme, EcoRI. You run the digested DNA on an agarose gel, and obtain the following fragments (all sizes are in kilobases):
e) Draw a restriction map of the DK15 DNA that is consistent with all the data provided. Be sure your map includes: 1) the location of all restriction sites; 2) the distances between restriction sites; 3) the total DNA size.
Anda mungkin juga menyukai
- CH 10 Test Bank For Essential Cell Biology 3rd Edition AlbertsDokumen35 halamanCH 10 Test Bank For Essential Cell Biology 3rd Edition AlbertsRokia GhariebBelum ada peringkat
- Error-Correction on Non-Standard Communication ChannelsDari EverandError-Correction on Non-Standard Communication ChannelsBelum ada peringkat
- Essential Cell Biology 3rd Edition Bruce Alberts Test BankDokumen28 halamanEssential Cell Biology 3rd Edition Bruce Alberts Test Banknhiamandat4uvr100% (27)
- It's In Your DNA! What Is DNA? - Biology Book 6th Grade | Children's Biology BooksDari EverandIt's In Your DNA! What Is DNA? - Biology Book 6th Grade | Children's Biology BooksBelum ada peringkat
- Essential Cell Biology 3rd Edition Bruce Alberts Test Bank Full Chapter PDFDokumen49 halamanEssential Cell Biology 3rd Edition Bruce Alberts Test Bank Full Chapter PDFoglepogy5kobgk100% (13)
- An Introduction to Stochastic Modeling, Student Solutions Manual (e-only)Dari EverandAn Introduction to Stochastic Modeling, Student Solutions Manual (e-only)Belum ada peringkat
- Practice ExamDokumen9 halamanPractice ExamVanandi100% (1)
- Strategy For Solving Restriction Digestion ProblemsDokumen2 halamanStrategy For Solving Restriction Digestion ProblemsPranav tembe100% (4)
- CH 5 Test Bank For Essential Cell Biology 3rd Edition AlbertsDokumen36 halamanCH 5 Test Bank For Essential Cell Biology 3rd Edition AlbertsRokia GhariebBelum ada peringkat
- Biosc1940 Molecular Biology Midterm Exam 1: Multiple Choice (2 Pts Each) - Circle One Answer For EachDokumen10 halamanBiosc1940 Molecular Biology Midterm Exam 1: Multiple Choice (2 Pts Each) - Circle One Answer For EachMohamed MounirBelum ada peringkat
- CH 3 - ProblemsDokumen6 halamanCH 3 - ProblemsKhris Griffis100% (5)
- Bio 140 Exam 4 Fall 2015Dokumen7 halamanBio 140 Exam 4 Fall 2015lp_blackoutBelum ada peringkat
- FBMDokumen7 halamanFBMnicpreBelum ada peringkat
- Experiment 7 - Plasmid Dna IsolationDokumen4 halamanExperiment 7 - Plasmid Dna IsolationDilay RıdvanBelum ada peringkat
- Numericals On DNA TopologyDokumen5 halamanNumericals On DNA TopologySadab KhanBelum ada peringkat
- DNA Databases - Evaluative Activity - José Ricardo Ramírez y Juan Diego RojasDokumen9 halamanDNA Databases - Evaluative Activity - José Ricardo Ramírez y Juan Diego Rojasjuan diegoBelum ada peringkat
- Solutions To 7.012 Problem Set 5: 5' G 3' Cleavage by Bamhi Leaves A 5' Overhang 3' Cctag 5'Dokumen5 halamanSolutions To 7.012 Problem Set 5: 5' G 3' Cleavage by Bamhi Leaves A 5' Overhang 3' Cctag 5'Ankita KandalkarBelum ada peringkat
- FHSC1234 Tutorial 8S 202210Dokumen5 halamanFHSC1234 Tutorial 8S 202210Zhi YongBelum ada peringkat
- Genetics Final Exam Study GuideDokumen24 halamanGenetics Final Exam Study GuideJesse Jae0% (1)
- 3b Manual AnswersDokumen2 halaman3b Manual AnswersImDman AsBelum ada peringkat
- RDT QuestionsDokumen5 halamanRDT QuestionsKedar SharmaBelum ada peringkat
- The Dna StructureDokumen28 halamanThe Dna Structuremark gonzalesBelum ada peringkat
- 22 Restriction Enzyme SimDokumen6 halaman22 Restriction Enzyme SimS. Spencer0% (1)
- BioBlitz 2022 FinalDokumen14 halamanBioBlitz 2022 FinalJamesBelum ada peringkat
- DNA Replication - PPT 1Dokumen48 halamanDNA Replication - PPT 1Keira MendozaBelum ada peringkat
- BS20001 4th Test - 18th November 2020Dokumen3 halamanBS20001 4th Test - 18th November 2020Hrushi RiyanBelum ada peringkat
- BS2009 Practicals 2008Dokumen4 halamanBS2009 Practicals 2008Bi AnhBelum ada peringkat
- Restriction Enzymes ActivityDokumen7 halamanRestriction Enzymes ActivityDoree GlantzBelum ada peringkat
- Druid Dracula PPT Case StudyDokumen28 halamanDruid Dracula PPT Case StudyKory TurnerBelum ada peringkat
- 5 Lesson8 Worksheets WithAnswersDokumen14 halaman5 Lesson8 Worksheets WithAnswersRa MilBelum ada peringkat
- Biol 201 - Exam 2 - KeyDokumen7 halamanBiol 201 - Exam 2 - KeyLama ZeinBelum ada peringkat
- Dna Computing: Presented By:-Anupam Kumar S-7, IT A'Dokumen54 halamanDna Computing: Presented By:-Anupam Kumar S-7, IT A'arya_accentBelum ada peringkat
- Molecular Biology of The Cell 5Th Edition Alberts Test Bank Full Chapter PDFDokumen34 halamanMolecular Biology of The Cell 5Th Edition Alberts Test Bank Full Chapter PDFrorybridgetewe100% (8)
- Restriction LabDokumen8 halamanRestriction Labtworedpartyhats100% (3)
- Unit-Ii Asm 2022-23Dokumen33 halamanUnit-Ii Asm 2022-23AnshnuBelum ada peringkat
- Molecular Biology of The Cell 5th Edition Alberts Test BankDokumen13 halamanMolecular Biology of The Cell 5th Edition Alberts Test Bankodettedieupmx23m100% (31)
- CArter Mostrom - Student Lab (These Pages Are Compact)Dokumen26 halamanCArter Mostrom - Student Lab (These Pages Are Compact)Carter MostromBelum ada peringkat
- Cut and PasteDokumen2 halamanCut and PasteKelsey PerreaultBelum ada peringkat
- Lab #2Dokumen8 halamanLab #2yejiBelum ada peringkat
- Production of Recombinant MoleculesDokumen3 halamanProduction of Recombinant MoleculesGuruKPOBelum ada peringkat
- Dna Computing: Deepthi Bollu CSE 497:computational Issues in Molecular Biology Professor-Dr. Lopresti April 13, 2004Dokumen54 halamanDna Computing: Deepthi Bollu CSE 497:computational Issues in Molecular Biology Professor-Dr. Lopresti April 13, 2004Kapil BhagavatulaBelum ada peringkat
- FGIKReplicationTeacherKey6!18!15 CompressedDokumen16 halamanFGIKReplicationTeacherKey6!18!15 CompressedSantiago Teixeira100% (1)
- Class 12 Bio PaperDokumen2 halamanClass 12 Bio PaperVector AcademyBelum ada peringkat
- Chapter 11 Test BankDokumen11 halamanChapter 11 Test Bankanon_747148947Belum ada peringkat
- HBS 131 ResponseSheetDokumen3 halamanHBS 131 ResponseSheetDelilah YacoubBelum ada peringkat
- L3-GeneTech DNA SequencingDokumen83 halamanL3-GeneTech DNA SequencingSwayamBelum ada peringkat
- Ex2 AnsDokumen2 halamanEx2 AnsShaheen TariqBelum ada peringkat
- Chip-Chip: Data, Model, and Analysis: Ming Zheng, Leah O. Barrera, Bing Ren, and Ying Nian WuDokumen10 halamanChip-Chip: Data, Model, and Analysis: Ming Zheng, Leah O. Barrera, Bing Ren, and Ying Nian WuGautam JhaBelum ada peringkat
- Exercises: The Site at Which It Cuts DnaDokumen3 halamanExercises: The Site at Which It Cuts DnaSubhadip MurmuBelum ada peringkat
- Biotechnology Principles and ProcessDokumen2 halamanBiotechnology Principles and ProcessMayank Kumar Singh.Belum ada peringkat
- 1.3 Reading GuideDokumen7 halaman1.3 Reading GuideriyaBelum ada peringkat
- 2019 Evomics Reference FreeDokumen118 halaman2019 Evomics Reference FreedrcyberBelum ada peringkat
- CSIR JRF Paper II (Subjective) Model Question1Dokumen1 halamanCSIR JRF Paper II (Subjective) Model Question1kuldip.biotechBelum ada peringkat
- Dna FingerprintingDokumen66 halamanDna Fingerprintingharun al hassan khanBelum ada peringkat
- PCB3103C Exam 2 (Take-Home)Dokumen5 halamanPCB3103C Exam 2 (Take-Home)Pmorris1898Belum ada peringkat
- Final MolecularDokumen13 halamanFinal Molecularpharmadr2024Belum ada peringkat
- 03Q PDFDokumen2 halaman03Q PDFFCiênciasBelum ada peringkat
- Beyond Silicon Computing: Jigar Purohit (03ce117) Gunjan Dholakiya (03ce116)Dokumen21 halamanBeyond Silicon Computing: Jigar Purohit (03ce117) Gunjan Dholakiya (03ce116)Disneyland27100% (6)
- Dna Computing: Using Dna To Solve Computational ProblemsDokumen12 halamanDna Computing: Using Dna To Solve Computational ProblemsJBelum ada peringkat
- 8 - Secondary Metabolites - TERPENOIDS ALKALOIDS PHENOLICS PDFDokumen82 halaman8 - Secondary Metabolites - TERPENOIDS ALKALOIDS PHENOLICS PDFFCiênciasBelum ada peringkat
- 01Q PDFDokumen1 halaman01Q PDFFCiênciasBelum ada peringkat
- Amber 15Dokumen883 halamanAmber 15FCiênciasBelum ada peringkat
- 03Q PDFDokumen2 halaman03Q PDFFCiênciasBelum ada peringkat
- 7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer KeyDokumen10 halaman7.02 Recombinant DNA Methods Spring 2005 Exam Study Questions Answer KeyFCiênciasBelum ada peringkat
- 1ºteste 2013 2014 Catarina Cunha PDFDokumen4 halaman1ºteste 2013 2014 Catarina Cunha PDFFCiênciasBelum ada peringkat
- Exercício Mutagénese Sítio Dirigida - Questões PDFDokumen25 halamanExercício Mutagénese Sítio Dirigida - Questões PDFFCiênciasBelum ada peringkat
- Aula 6: Bioquimica IIDokumen21 halamanAula 6: Bioquimica IIFCiênciasBelum ada peringkat
- Alcohol - Metab p450 PDFDokumen17 halamanAlcohol - Metab p450 PDFFCiênciasBelum ada peringkat
- Review: S. E. Borggreve, R. de Vries and R. P. F. DullaartDokumen19 halamanReview: S. E. Borggreve, R. de Vries and R. P. F. DullaartFCiênciasBelum ada peringkat
- LipidogramaDokumen1 halamanLipidogramaFCiênciasBelum ada peringkat
- Aula071314 PDFDokumen33 halamanAula071314 PDFFCiênciasBelum ada peringkat
- S1744309110011589 PDFDokumen5 halamanS1744309110011589 PDFFCiênciasBelum ada peringkat
- 08 Recombination PDFDokumen17 halaman08 Recombination PDFFCiênciasBelum ada peringkat
- 08 Recombination PDFDokumen15 halaman08 Recombination PDFFCiênciasBelum ada peringkat
- Return of the God Hypothesis: Three Scientific Discoveries That Reveal the Mind Behind the UniverseDari EverandReturn of the God Hypothesis: Three Scientific Discoveries That Reveal the Mind Behind the UniversePenilaian: 4.5 dari 5 bintang4.5/5 (52)
- A Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsDari EverandA Brief History of Intelligence: Evolution, AI, and the Five Breakthroughs That Made Our BrainsPenilaian: 4.5 dari 5 bintang4.5/5 (6)
- 10% Human: How Your Body's Microbes Hold the Key to Health and HappinessDari Everand10% Human: How Your Body's Microbes Hold the Key to Health and HappinessPenilaian: 4 dari 5 bintang4/5 (33)
- Why We Die: The New Science of Aging and the Quest for ImmortalityDari EverandWhy We Die: The New Science of Aging and the Quest for ImmortalityPenilaian: 4 dari 5 bintang4/5 (5)
- When the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisDari EverandWhen the Body Says No by Gabor Maté: Key Takeaways, Summary & AnalysisPenilaian: 3.5 dari 5 bintang3.5/5 (2)
- The Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RaceDari EverandThe Molecule of More: How a Single Chemical in Your Brain Drives Love, Sex, and Creativity--and Will Determine the Fate of the Human RacePenilaian: 4.5 dari 5 bintang4.5/5 (517)
- The Rise and Fall of the Dinosaurs: A New History of a Lost WorldDari EverandThe Rise and Fall of the Dinosaurs: A New History of a Lost WorldPenilaian: 4 dari 5 bintang4/5 (597)
- Masterminds: Genius, DNA, and the Quest to Rewrite LifeDari EverandMasterminds: Genius, DNA, and the Quest to Rewrite LifeBelum ada peringkat
- Gut: the new and revised Sunday Times bestsellerDari EverandGut: the new and revised Sunday Times bestsellerPenilaian: 4 dari 5 bintang4/5 (393)
- Tales from Both Sides of the Brain: A Life in NeuroscienceDari EverandTales from Both Sides of the Brain: A Life in NeurosciencePenilaian: 3 dari 5 bintang3/5 (18)
- Buddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomDari EverandBuddha's Brain: The Practical Neuroscience of Happiness, Love & WisdomPenilaian: 4 dari 5 bintang4/5 (216)
- Seven and a Half Lessons About the BrainDari EverandSeven and a Half Lessons About the BrainPenilaian: 4 dari 5 bintang4/5 (111)
- All That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesDari EverandAll That Remains: A Renowned Forensic Scientist on Death, Mortality, and Solving CrimesPenilaian: 4.5 dari 5 bintang4.5/5 (397)
- Undeniable: How Biology Confirms Our Intuition That Life Is DesignedDari EverandUndeniable: How Biology Confirms Our Intuition That Life Is DesignedPenilaian: 4 dari 5 bintang4/5 (11)
- Good Without God: What a Billion Nonreligious People Do BelieveDari EverandGood Without God: What a Billion Nonreligious People Do BelievePenilaian: 4 dari 5 bintang4/5 (66)
- The Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionDari EverandThe Ancestor's Tale: A Pilgrimage to the Dawn of EvolutionPenilaian: 4 dari 5 bintang4/5 (812)
- Human: The Science Behind What Makes Your Brain UniqueDari EverandHuman: The Science Behind What Makes Your Brain UniquePenilaian: 3.5 dari 5 bintang3.5/5 (38)
- Lymph & Longevity: The Untapped Secret to HealthDari EverandLymph & Longevity: The Untapped Secret to HealthPenilaian: 4.5 dari 5 bintang4.5/5 (13)
- The Lives of Bees: The Untold Story of the Honey Bee in the WildDari EverandThe Lives of Bees: The Untold Story of the Honey Bee in the WildPenilaian: 4.5 dari 5 bintang4.5/5 (44)
- Who's in Charge?: Free Will and the Science of the BrainDari EverandWho's in Charge?: Free Will and the Science of the BrainPenilaian: 4 dari 5 bintang4/5 (65)
- Darwin's Doubt: The Explosive Origin of Animal Life and the Case for Intelligent DesignDari EverandDarwin's Doubt: The Explosive Origin of Animal Life and the Case for Intelligent DesignPenilaian: 4.5 dari 5 bintang4.5/5 (39)
- Fast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperDari EverandFast Asleep: Improve Brain Function, Lose Weight, Boost Your Mood, Reduce Stress, and Become a Better SleeperPenilaian: 4.5 dari 5 bintang4.5/5 (16)
- The Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindDari EverandThe Consciousness Instinct: Unraveling the Mystery of How the Brain Makes the MindPenilaian: 4.5 dari 5 bintang4.5/5 (93)
- Gut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)Dari EverandGut: The Inside Story of Our Body's Most Underrated Organ (Revised Edition)Penilaian: 4 dari 5 bintang4/5 (411)
- The Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorDari EverandThe Other Side of Normal: How Biology Is Providing the Clues to Unlock the Secrets of Normal and Abnormal BehaviorBelum ada peringkat