Appendix Scripts
Diunggah oleh
kensaiiDeskripsi Asli:
Hak Cipta
Format Tersedia
Bagikan dokumen Ini
Apakah menurut Anda dokumen ini bermanfaat?
Apakah konten ini tidak pantas?
Laporkan Dokumen IniHak Cipta:
Format Tersedia
Appendix Scripts
Diunggah oleh
kensaiiHak Cipta:
Format Tersedia
Final Report: Appendix
Damiano Milani
In this appendix some of the scripts used in the Report are attached.
1 Shell script
This script, in the sh language, manages the instructions to launch the LAMMPS simula-
tions. It automatically loads the input les for each type of simulations and extract the
numerical data from the output.
#!/bin/bash
#
# FINAL REPORT - Nanomechanics
# Shell script
#
# Damiano Milani
#
#source les path
source_path="/home/dami/nanomechanics/final_report"
#LAMMPS path
lammps_path="/home/dami/.lammps/src/lmp_openmpi"
clear
echo -e "NANOMECHANICS - FINAL REPORT
Damiano Milani\n
Choose the script:
1) Cu Nanorod (1D)
2) Cu Film (2D)
3) Cu Bulk (3D)
4) Cu Indentation (2D)
5) Elastic parameters
6) Exit\n"
read task;
echo -e "\nChoose the revision:"
read rev;
clear
case $task in
1)
## 1 NANOROD
echo -e "Task 1 - Cu 1D Nanorod\n"
# goto source le path
cd $source_path/Cu-1D/$rev/
step_e="101"
line_t="9,109p;132,932p;955,1055p"
;;
1
2)
## 2 FILM
clear
echo -e "Task 2 - Cu 2D Film\n"
cd $source_path/Cu-2D/$rev/
step_e="101"
line_t="9,109p;132,932p;955,1055p"
;;
3)
## 3 BULK
clear
echo -e "Task 3 - Cu 3D Bulk\n"
cd $source_path/Cu-3D/$rev/
step_e="201"
line_t="9,110p;133,934p;957,1058p"
;;
4)
## 4 INDENTATION
clear
echo -e "Task 4 - Cu Indentation\n"
cd $source_path/Cu-indent/$rev/
echo "running indent..."
#mpirun -np 2 $lammps_path < in.indent > out.indent
grep "Loop time" out.indent | grep [0-9]
echo -e "indent completed!\n\n"
# data visualization
less out.indent
# data extraction
cat out.indent | sed -n -e "232,433p;456,506p;529,579p" > data.indent
read
exit
;;
5)
## 5 ELASTIC CONSTANTS
clear
echo "Task 5 - Elastic constants, fcc Ni"
echo "running..."
cd $source_path/elastic
mpirun -np 2 $lammps_path < in.elastic > out.elastic
less out.elastic
tail -n 21 out.elastic > data.elastic
echo "completed!"
read
exit
;;
6) exit;;
esac;
## MAIN
# run lammps
echo "running equil..."
mpirun -np 2 $lammps_path < in.equil > out.equil
grep "Loop time" out.equil | grep [0-9]
echo -e "equil done!\n\n"
echo "running tension..."
mpirun -np 2 $lammps_path < in.tension > out.tension
grep "Loop time" out.tension | grep [0-9]
echo -e "tension done!\n\ntask completed!"
# visual output
less out.equil
less out.tension
# extract data
grep -A$step_e Step out.equil | grep [0-9] | tail --lines=$step_e > data.equil
cat out.tension | sed -n -e "$line_t" > data.tension
read
2 LAMMPS scripts
2.1 Equilibration script
In the following script the atom box is created (fcc copper), and then equilibrated mini-
mizing the energy at 300 K with NPT ensamble (note the EAM potential).
# in.equil
# REVISION 3
#
# Damiano Milani
#
# Cu nanorod equilibration
#
# eps=1.5 T=300
variable T equal 300.
# system set-up
log log.equil
units metal
dimension 3
boundary s s p
atom_style atomic
neighbor 0.3 bin
neigh_modify delay 5
# create geometry
lattice fcc 3.6145
region box block -3 3 -3 3 -10 10
create_box 1 box
create_atoms 1 box
# Cu EAM potential
pair_style eam
pair_coeff * * Cu_u6.eam
# minimize energy
thermo 100
thermo_style custom step temp etotal pe ke press pxx pyy pzz lx ly lz vol
dump 1 all atom 1000 dump.equil
fix min all box/relax z 1.
minimize 0. 0. 1000 10000
# dynamics setup
timestep 0.001
fix 1 all npt temp ${T} ${T} 0.200 z 1. 1. .6
# equilibrate the system at T = 300 K
variable T2 equal 2*${T}
velocity all create ${T2} 19529
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 98235
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 532518
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 251820
run 10000
write_restart restart.equil
2.2 Tension script
In this script the tensile stress is applied: the box is stretched until the maximum strain
set; then it is freely relaxed.
# in.tension
# REVISION 3
#
# Damiano Milani
#
# Cu nanorod tension
#
# eps=1.5 T=300
log log.nanorod
# nal z-strain
variable max_strain equal 1.5
# temperature
variable T equal 300.
# system set-up
units metal
dimension 3
boundary s s p
atom_style atomic
neighbor 0.3 bin
neigh_modify delay 5
# read equilibrated structure
read_restart restart.equil
# Cu EAM potential
pair_style eam
pair_coeff * * Cu_u6.eam
# outputs
thermo 100
thermo_style custom step temp etotal pe ke press pxx pyy pzz lx ly lz vol
dump 1 all atom 100 dump.tension
# equilibrate the system at the specied temperature
timestep 0.001
fix 1 all nvt temp ${T} ${T} .2
run 10000
# tension or compression test
fix 2 all deform 10 z scale ${max_strain}
run 80000
# release the load
unfix 1
unfix 2
fix 1 all npt temp ${T} ${T} .2 z 1. 1. .6
run 10000
2.3 Strain rate script
For changing the strain rate, the number of step and/or the lenght of the timesteps during
the deformation could be varied as shown below:
# in.tension
# REVISION 12
#
# Damiano Milani
#
# Cu nanorod tension
#
# eps=3 T=10 vel=1x
log log.nanorod
# nal z-strain
variable max_strain equal 1.5
# temperature
variable T equal 300.
# system set-up
units metal
dimension 3
boundary s s p
atom_style atomic
neighbor 0.3 bin
neigh_modify delay 5
# read equilibrated structure
read_restart restart.equil
# Cu EAM potential
pair_style eam
pair_coeff * * Cu_u6.eam
# outputs
thermo 400
thermo_style custom step temp etotal pe ke press pxx pyy pzz lx ly lz vol
dump 1 all atom 100 dump.tension
# equilibrate the system at the specied temperature
timestep 0.001
fix 1 all nvt temp ${T} ${T} .2
run 40000
# tension or compression test
fix 2 all deform 10 z scale ${max_strain}
run 320000
# release the load
unfix 1
unfix 2
fix 1 all npt temp ${T} ${T} .2 z 1. 1. .6
run 40000
2.4 Size script
For increasing the cross section of the nanorod, in the creation of the box the number of
cells is increased:
# in.equil
# REVISION 23
#
# Damiano Milani
#
# Cu nanorod equilibration
#
# eps=1.5 T=300 size=9a
variable T equal 300.
# system set-up
log log.equil
units metal
dimension 3
boundary s s p
atom_style atomic
neighbor 0.3 bin
neigh_modify delay 5
# create geometry
lattice fcc 3.6145
region box block -4.5 4.5 -4.5 4.5 -15 15
create_box 1 box
create_atoms 1 box
# Cu EAM potential
pair_style eam
pair_coeff * * Cu_u6.eam
# minimize energy
thermo 100
thermo_style custom step temp etotal pe ke press pxx pyy pzz lx ly lz vol
dump 1 all atom 1000 dump.equil
fix min all box/relax z 1.
minimize 0. 0. 1000 10000
# dynamics setup
timestep 0.001
fix 1 all npt temp ${T} ${T} 0.200 z 1. 1. .6
# equilibrate the system at T = 300 K
variable T2 equal 2*${T}
velocity all create ${T2} 19529
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 98235
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 532518
run 500
minimize 0. 0. 1000 10000
velocity all create ${T2} 251820
run 10000
write_restart restart.equil
3 MATLAB scripts
3.1 Main script
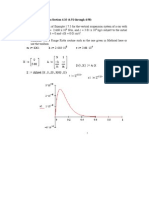
This script provides a complete data processing, for each of the topic dealt in the Report:
the temperature, the strain rate and the size effect. It plots the analyzed curves, and it
makes the interpolations to determine the mechanical parameters.
At the end there is also a part for calculating elastic parameters of copper (Reuss, Voigt,
Kroner model), using the results from the ELASTIC package in LAMMPS.
%% Nanomechanics - FINAL REPORT %%
% Damiano Milani
close all
clear all
clc
global colore
global titolo
global Step
global Temp
global TotEng
global PotEng
global KinEng
global Press
global Pxx
global Pyy
global Pzz
global Lx
global Ly
global Lz
global Volume
global Step_t
global Temp_t
global TotEng_t
global PotEng_t
global KinEng_t
global Press_t
global Pxx_t
global Pyy_t
global Pzz_t
global Lx_t
global Ly_t
global Lz_t
global Volume_t
% colorize vector
col=[brgcmykbrgcmykbrgcmykbbrgcmykbrgcmyk];
task= 2 ;
switch task
%% Task 1 - Nanorod - Temp variation
case 1
E=[];
s_y=[];
temperatures=[10,100,200,300,400,500];
for i=[1:6]
load_data(1,i);
epszz=(Lz_t-Lz_t(1))/Lz_t(1);
figure(1)
plot(epszz,-Pzz_t,col(i),linewidth,1.5)
[yield n]=max(abs(Pzz_t(1:300)));
[cf stat]=polyfit(epszz(1:n),-Pzz_t(1:n),1);
elastic=polyval(cf,epszz(1:n));
hold on
plot(epszz(1:n),elastic,k--,linewidth,1)
plot(epszz(1:n),yield,g-,linewidth,2)
xlabel(\epsilon)
ylabel(\sigma [MPa])
axis([0 0.51 -1000 +11000])
grid on
E=[E,cf(1)];
s_y=[s_y, yield];
figure(10)
hold on
plot(temperatures(i),E(i),[col(i) *],linewidth,4)
figure(11)
hold on
plot(temperatures(i),s_y(i),[col(i) *],linewidth,4)
end
E
s_y
figure(1)
legend(T=10 K,T=100 K,T=200 K,T=300 K,T=400 K,T=500 K)
figure(10)
plot([10,100,200,300,400,500],E,r--,linewidth,1)
xlabel(T [K])
ylabel(E [MPa])
axis([0 550 25000 +75000])
grid on
figure(11)
plot([10,100,200,300,400,500],s_y,g--,linewidth,1)
xlabel(T [K])
ylabel(\sigma_y [MPa])
axis([0 550 1000 +11000])
grid on
%% Task 2 - Nanorod - Strain rate
case 2
E=[];
s_y=[];
vel=[1e11,5e10,2.5e10,6.25e9];
for i=[10:13]
load_data(1,i);
epszz=(Lz_t-Lz_t(1))/Lz_t(1);
figure(2)
hold on
plot(Lz_t,col(i))
figure(1)
plot(epszz,-Pzz_t,col(i),linewidth,1)
[yield n]=max(abs(Pzz_t));
[cf stat]=polyfit(epszz(1:n),-Pzz_t(1:n),1);
elastic=polyval(cf,epszz(1:n));
hold on
%plot(epszz(1:n),elastic,k)
xlabel(\epsilon)
ylabel(\sigma [MPa])
grid on
axis([0 2.1 -1000 +16000])
legend(1e11 s-1,5e10 s-1,2.5e10 s-1,6.25e9 s-1);
E=[E,cf(1)];
s_y=[s_y, yield];
figure(10)
hold on
plot(vel(i-9),cf(1),[col(i) *],linewidth,4)
figure(11)
hold on
plot(vel(i-9),s_y(i-9),[col(i) *],linewidth,4)
end
E
s_y
figure(10)
hold on
semilogx(vel,E,r--,linewidth,1)
xlabel(d\epsilon/dt [s-1])
ylabel(\sigma [MPa])
axis([1e9 1.1e11 72e3 80e3])
grid on
figure(11)
hold on
semilogx(vel,s_y,g--,linewidth,1)
xlabel(d\epsilon/dt [s-1])
ylabel(\sigma_y [MPa])
axis([1e9 1.1e11 .9e4 1.7e4])
grid on
%% Task 3 - Nanorod - Size effect
case 3
E=[];
s_y=[];
side=[];
for i=[20:23]
load_data(1,i);
epszz=(Lz_t-Lz_t(1))/Lz_t(1);
figure(1)
plot(epszz,-Pzz_t,col(i-5),linewidth,1)
[yield n]=max(abs(Pzz_t));
[cf stat]=polyfit(epszz(1:n),-Pzz_t(1:n),1);
elastic=polyval(cf,epszz(1:n));
hold on
%plot(epszz(1:n),elastic,k)
xlabel(\epsilon)
ylabel(\sigma [MPa])
grid on
axis([0 .51 -1000 +12000])
legend(4ax4a,5ax5a,7ax7a,9ax9a);
figure(20)
plot(epszz)
E=[E,cf(1)];
s_y=[s_y, yield];
side=[side Lx_t(1)]
figure(10)
hold on
plot(side(i-19),cf(1),[col(i-5) *],linewidth,4)
figure(11)
hold on
plot(side(i-19),s_y(i-19),[col(i-5) *],linewidth,4)
end
E
s_y
figure(10)
hold on
plot(side,E,r--,linewidth,1)
xlabel(L_x [A])
ylabel(\sigma [MPa])
%axis([1e9 1.1e11 72e3 80e3])
grid on
figure(11)
hold on
plot(side,s_y,g--,linewidth,1)
xlabel(L_x [A])
ylabel(\sigma_y [MPa])
%axis([1e9 1.1e11 .9e4 1.7e4])
grid on
%% Elastic Parameters
case 6
% voigt
clear all
a=168;
b=123;
c=78;
E=((a - b + 3*c)*(a + 2*b))/(2*a + 3*b + c)
v=(a + 4*b - 2*c)/(2*(2*a + 3*b + c))
G=E/2/(1+v)
K=E/3/(1-2*v)
% kroner
clear all
a=168;
b=123;
c=78;
p=(a+b)/(a-b)/(a+2*b);
q=-b/(a-b)/(a+2*b);
r=1/c;
mu1=0.5*(a-b);
mu2=c;
K=(a+2*b)/3
g1=(9*K+4*mu1)/8;
g2=-mu2*(3*K+12*mu1)/8;
g3=-3*K*mu1*mu2/4;
syms G;
f=G3+f1*G2+f2*G+f3;
f=subs(f,f1,g1);
f=subs(f,f2,g2);
f=subs(f,f3,g3);
res=solve(f,G);
re1=eval(res(1))
E=(9*K*re1)/(3*K+re1)
v=(3*K-2*re1)/2/(3*K+re1)
%reuss
clear all
a=168;
b=123;
c=78;
p=(a+b)/(a-b)/(a+2*b);
q=-b/(a-b)/(a+2*b);
r=1/c;
for gamma=[0,1/4,1/3] %100 110 111 directions
s0=p-q-r/2;
E=1/(p-2*s0*gamma)
end
end
%% finishing
tilefigs
3.2 Load script
The following is the custom function for loading data from output le of LAMMPS, and
to allow to use them in MATLAB as normal variables.
function load_data( choice, rev )
%LOAD_DATA load data from LAMMPS output file
% choice is the task folder
% rev is the revision subfolder
global colore
global titolo
global Step
global Temp
global TotEng
global PotEng
global KinEng
global Press
global Pxx
global Pyy
global Pzz
global Lx
global Ly
global Lz
global Volume
global Step_e
global Temp_e
global TotEng_e
global PotEng_e
global KinEng_e
global Press_e
global Pxx_e
global Pyy_e
global Pzz_e
global Lx_e
global Ly_e
global Lz_e
global Volume_e
global Step_t
global Temp_t
global TotEng_t
global PotEng_t
global KinEng_t
global Press_t
global Pxx_t
global Pyy_t
global Pzz_t
global Lx_t
global Ly_t
global Lz_t
global Volume_t
%task directory and labels
switch choice
case 1
cd Cu-1D
colore=b;
titolo=Cu Nanorod;
case 2
cd Cu-2D
colore=c;
titolo=Cu Film;
case 3
cd Cu-3D
colore=g;
titolo=Cu Bulk;
case 4
cd Cu-indent
colore=r;
titolo=Cu indentation;
end
%revision dir
rev=num2str(rev);
cd(rev)
% load data file
equil=load(data.equil);
tension=load(data.tension);
% DATA STRUCTURE:
% Step Temp TotEng PotEng KinEng Press Pxx Pyy Pzz Lx Ly Lz Volume
%equilibrium data
Step_e = equil(:,1);
Temp_e = equil(:,2); % kelvin
TotEng_e = equil(:,3); % eV
PotEng_e = equil(:,4);
KinEng_e = equil(:,5);
Press_e = equil(:,6)/10; % bar->MPa
Pxx_e = equil(:,7)/10;
Pyy_e = equil(:,8)/10;
Pzz_e = equil(:,9)/10;
Lx_e = equil(:,10); % angstrom
Ly_e = equil(:,11);
Lz_e = equil(:,12);
Volume_e = equil(:,13);
%tension data
Step_t = tension(:,1);
Temp_t = tension(:,2);
TotEng_t = tension(:,3);
PotEng_t = tension(:,4);
KinEng_t = tension(:,5);
Press_t = tension(:,6)/10;
Pxx_t = tension(:,7)/10;
Pyy_t = tension(:,8)/10;
Pzz_t = tension(:,9)/10;
Lx_t = tension(:,10);
Ly_t = tension(:,11);
Lz_t = tension(:,12);
Volume_t = tension(:,13);
%complete data
Step = [Step_e;Step_t];
Temp = [Temp_e;Temp_t];
TotEng = [TotEng_e;TotEng_t];
PotEng = [PotEng_e;PotEng_t];
KinEng = [KinEng_e;KinEng_t];
Press = [Press_e;Press_t];
Pxx = [Pxx_e;Pxx_t];
Pyy = [Pyy_e;Pyy_t];
Pzz = [Pzz_e;Pzz_t];
Lx = [Lx_e;Lx_t];
Ly = [Ly_e;Ly_t];
Lz = [Lz_e;Lz_t];
Volume = [Volume_e;Volume_t];
%main directory
cd ../..
end
Anda mungkin juga menyukai
- LammpsDokumen21 halamanLammpsFernandoBelum ada peringkat
- 3: Getting Started With LAMMPS: Steve Plimpton, Sjplimp@sandia - GovDokumen75 halaman3: Getting Started With LAMMPS: Steve Plimpton, Sjplimp@sandia - GovYeasir MahmudBelum ada peringkat
- Silica Glass Input FileDokumen1 halamanSilica Glass Input FileGiselle CarolinaBelum ada peringkat
- LAMMPSDokumen92 halamanLAMMPSdaniyalhassan2789Belum ada peringkat
- Lammps Ex. and ExplanationDokumen6 halamanLammps Ex. and Explanation17029 Ankon SahaBelum ada peringkat
- TMPKDokumen11 halamanTMPKRuswanto RiswanBelum ada peringkat
- Mechanical Vibration Lab ReportDokumen7 halamanMechanical Vibration Lab ReportChris NichollsBelum ada peringkat
- Uniaxial Tension - EVOCDDokumen7 halamanUniaxial Tension - EVOCDanon_637246413Belum ada peringkat
- Tutorial 1Dokumen11 halamanTutorial 1Witold SzejgisBelum ada peringkat
- Mumps TutorialDokumen8 halamanMumps TutorialShashank KaushalBelum ada peringkat
- OOP Lab 1Dokumen8 halamanOOP Lab 1Mohsin Ali0% (1)
- QMMM Tutorial Ws10 11Dokumen36 halamanQMMM Tutorial Ws10 11weilaiBelum ada peringkat
- Steve Plimpton Sandia National Labs Sjplimp@sandia - GovDokumen77 halamanSteve Plimpton Sandia National Labs Sjplimp@sandia - GovGeorge Michael Alvarado LopezBelum ada peringkat
- Aquino Lab04Dokumen18 halamanAquino Lab04Ai RahBelum ada peringkat
- LoveresLabExercise05Group06CAECHE51S1 PDFDokumen74 halamanLoveresLabExercise05Group06CAECHE51S1 PDFShania LoveresBelum ada peringkat
- Axi Symmetric and Conjugate Heat Transfer Problems - Validation of OpenFoam CodesDokumen17 halamanAxi Symmetric and Conjugate Heat Transfer Problems - Validation of OpenFoam CodesVijay KumarBelum ada peringkat
- Metnum Ex1Dokumen3 halamanMetnum Ex1baig79Belum ada peringkat
- Chapter 2: Objects and Primitive Data: SolutionsDokumen13 halamanChapter 2: Objects and Primitive Data: SolutionsEric Martin100% (1)
- Damping SystemDokumen6 halamanDamping SystemEro DoppleganggerBelum ada peringkat
- Examsimulation1 SolutionsDokumen9 halamanExamsimulation1 SolutionsardizzzBelum ada peringkat
- Ns2 Streaming Implementations Tutorial VCarrascalDokumen11 halamanNs2 Streaming Implementations Tutorial VCarrascalnoormohd99Belum ada peringkat
- Howto LiggghtsDokumen4 halamanHowto Liggghtsestebang79Belum ada peringkat
- HSPICEGuideDokumen45 halamanHSPICEGuideVinny LamBelum ada peringkat
- LabExercise 6 - Single Non-Linear EquationsDokumen32 halamanLabExercise 6 - Single Non-Linear EquationsQueenie Rose PercilBelum ada peringkat
- GoodDokumen44 halamanGoodDeni haryadiBelum ada peringkat
- NX Nastran PSD RMS Von Mises Stress Calculator Rev-1Dokumen5 halamanNX Nastran PSD RMS Von Mises Stress Calculator Rev-1GustavoYippeekayyayGusBelum ada peringkat
- Homework Assignment 3 Homework Assignment 3Dokumen10 halamanHomework Assignment 3 Homework Assignment 3Ido AkovBelum ada peringkat
- Roms NotesDokumen8 halamanRoms NotesAlexis QuirozBelum ada peringkat
- Guide CubicDokumen21 halamanGuide CubicFarhat Ullah Marwat PhysicistBelum ada peringkat
- How To Configure ACCDokumen6 halamanHow To Configure ACCalive2flirtBelum ada peringkat
- Quick Guide To Using Silvaco SUPREM.: Getting StartedDokumen9 halamanQuick Guide To Using Silvaco SUPREM.: Getting Startedapurva guptaBelum ada peringkat
- 1o9u.pdb (Renum - 1, Water & Ligand Remove) : 1. Extract The Residues Sequence by Using The Following ScriptDokumen6 halaman1o9u.pdb (Renum - 1, Water & Ligand Remove) : 1. Extract The Residues Sequence by Using The Following Scriptazhagar_ssBelum ada peringkat
- Ansys Interfacing With VC&FortranDokumen10 halamanAnsys Interfacing With VC&FortranBing HanBelum ada peringkat
- A Simple Example of LammpsDokumen8 halamanA Simple Example of LammpsLoekan JoeBelum ada peringkat
- Advanced Linux: Exercises: 0 Download and Unpack The Exercise Files (Do That First Time Only)Dokumen6 halamanAdvanced Linux: Exercises: 0 Download and Unpack The Exercise Files (Do That First Time Only)Niran SpiritBelum ada peringkat
- Thermo Calc Console ExamplesDokumen523 halamanThermo Calc Console ExamplesKarthi KeyanBelum ada peringkat
- SpectreRF Simulation TutorialDokumen110 halamanSpectreRF Simulation TutorialHaoWangBelum ada peringkat
- Datastage Realtime SenariosDokumen23 halamanDatastage Realtime SenariosArc100% (1)
- Implement Turbulence ModelDokumen12 halamanImplement Turbulence Modelnatuan74Belum ada peringkat
- Project 1Dokumen18 halamanProject 1soobashBelum ada peringkat
- Channel Tut v1Dokumen11 halamanChannel Tut v1Umair IsmailBelum ada peringkat
- Tutorial How To Check Your Grammar and DfaDokumen6 halamanTutorial How To Check Your Grammar and DfakavirangaBelum ada peringkat
- 6 Lab ExerciseDokumen12 halaman6 Lab ExerciseMia Beatrice Benavidez Mawili100% (1)
- SolSec 4 - 10 PDFDokumen11 halamanSolSec 4 - 10 PDFHKTBelum ada peringkat
- Matlab CodeDokumen85 halamanMatlab CodeBeny Abdou100% (2)
- Preparing Incremental Driver InputDokumen6 halamanPreparing Incremental Driver InputLuis Felipe Prada SarmientoBelum ada peringkat
- Fx-82sx 220plus Etc enDokumen2 halamanFx-82sx 220plus Etc enMegan Graciela IceMintzBelum ada peringkat
- Nas113 Composites Workbook 2Dokumen24 halamanNas113 Composites Workbook 2selva1975Belum ada peringkat
- Isatel MDS21 Clamp SW Operation ManualDokumen18 halamanIsatel MDS21 Clamp SW Operation ManualMarco Vinicio BazzottiBelum ada peringkat
- Wed 4 VerdiDokumen20 halamanWed 4 VerdiyapiBelum ada peringkat
- Exp - 08 Flight86Dokumen6 halamanExp - 08 Flight86Muhammad SaqibBelum ada peringkat
- Building A Reusable LAMMPS Script Library: Craig Tenney Edward MaginnDokumen24 halamanBuilding A Reusable LAMMPS Script Library: Craig Tenney Edward MaginnSwati VermaBelum ada peringkat
- 07 VXVM VXDMP (Dynamic Multi Pathing)Dokumen5 halaman07 VXVM VXDMP (Dynamic Multi Pathing)Sanjay KumarBelum ada peringkat
- Atom-4 2 0Dokumen17 halamanAtom-4 2 0Elias BritoBelum ada peringkat
- Windsim ManualDokumen8 halamanWindsim ManualNíal Otein ZemógBelum ada peringkat
- Wed 7 MargineDokumen27 halamanWed 7 Margineshubham patelBelum ada peringkat
- Build Model FastDokumen39 halamanBuild Model Fastluis BedrinanaBelum ada peringkat
- Projects With Microcontrollers And PICCDari EverandProjects With Microcontrollers And PICCPenilaian: 5 dari 5 bintang5/5 (1)
- Advanced Opensees Algorithms, Volume 1: Probability Analysis Of High Pier Cable-Stayed Bridge Under Multiple-Support Excitations, And LiquefactionDari EverandAdvanced Opensees Algorithms, Volume 1: Probability Analysis Of High Pier Cable-Stayed Bridge Under Multiple-Support Excitations, And LiquefactionBelum ada peringkat
- Sample Template For International Journal of Multiphase FlowDokumen1 halamanSample Template For International Journal of Multiphase FlowkensaiiBelum ada peringkat
- Sample Template For International Journal of Multiphase Flow - 5Dokumen1 halamanSample Template For International Journal of Multiphase Flow - 5kensaiiBelum ada peringkat
- Variational AutoencoderDokumen31 halamanVariational AutoencoderkensaiiBelum ada peringkat
- Sample Template For International Journal of Multiphase Flow - 2Dokumen1 halamanSample Template For International Journal of Multiphase Flow - 2kensaiiBelum ada peringkat
- Handson IVDokumen26 halamanHandson IVkensaiiBelum ada peringkat
- Italy Kappa Viscosity Mar14Dokumen47 halamanItaly Kappa Viscosity Mar14kensaiiBelum ada peringkat
- ShapeNet: An Information-Rich 3D Model RepositoryDokumen11 halamanShapeNet: An Information-Rich 3D Model RepositorykensaiiBelum ada peringkat
- 06 RNC NancyDokumen110 halaman06 RNC NancykensaiiBelum ada peringkat
- Lévy Processes and Their CharacteristicsDokumen23 halamanLévy Processes and Their CharacteristicskensaiiBelum ada peringkat
- JSM2016 ProgramBookDokumen311 halamanJSM2016 ProgramBookkensaiiBelum ada peringkat
- Super Quick Introduction To MPIDokumen32 halamanSuper Quick Introduction To MPIkensaiiBelum ada peringkat
- A Method For The Approximation of Functions Defined by Formal Series ExpansionsDokumen13 halamanA Method For The Approximation of Functions Defined by Formal Series ExpansionskensaiiBelum ada peringkat
- Green TheoremDokumen18 halamanGreen TheoremkensaiiBelum ada peringkat
- 05 CCA Kyoto2Dokumen47 halaman05 CCA Kyoto2kensaiiBelum ada peringkat
- DCS A-10C Keyboard LayoutDokumen7 halamanDCS A-10C Keyboard LayoutMichiel Erasmus100% (2)
- Career Planning Resources - 2021-22Dokumen1 halamanCareer Planning Resources - 2021-22Jessica NulphBelum ada peringkat
- Nurshath Dulal Wiki, Age, Photos, Height, Family, Net WorthDokumen1 halamanNurshath Dulal Wiki, Age, Photos, Height, Family, Net Worthraf0707066969Belum ada peringkat
- Business Law NMIMSDokumen10 halamanBusiness Law NMIMSprivate lessons0% (1)
- Form 15: Name of Member 1Dokumen2 halamanForm 15: Name of Member 1FSADFBelum ada peringkat
- The Ultimate C - C - TS4CO - 1909 - SAP Certified Application Associate - SAP S/4HANA For Management Accounting Associates (SAP S/4HANA 1909)Dokumen2 halamanThe Ultimate C - C - TS4CO - 1909 - SAP Certified Application Associate - SAP S/4HANA For Management Accounting Associates (SAP S/4HANA 1909)KirstingBelum ada peringkat
- Learning Activity Sheets (LAS) (For TLE/TVL CSS NCII)Dokumen6 halamanLearning Activity Sheets (LAS) (For TLE/TVL CSS NCII)Jåy-ž ShìzhènBelum ada peringkat
- CC-Link Function Manual: Motoman NX100 ControllerDokumen47 halamanCC-Link Function Manual: Motoman NX100 ControllersunhuynhBelum ada peringkat
- Mark Scheme End of Year Exam: EDPM Paper 2 Question 1 (Copy Test) Total 10 Marks AccuracyDokumen6 halamanMark Scheme End of Year Exam: EDPM Paper 2 Question 1 (Copy Test) Total 10 Marks AccuracyJanicSmithBelum ada peringkat
- Chapter FiveDokumen88 halamanChapter FiveErmias MesfinBelum ada peringkat
- Tutorial 12 13 14 BBBL2023Dokumen16 halamanTutorial 12 13 14 BBBL2023YANG YUN RUIBelum ada peringkat
- Elwave ManualDokumen109 halamanElwave ManualNo Name100% (1)
- ICT X Practice QuestionsDokumen3 halamanICT X Practice QuestionsYash KohaleBelum ada peringkat
- Core Java Material 2Dokumen199 halamanCore Java Material 2raamsgoluguriBelum ada peringkat
- Tally Prime Course Account and IntroDokumen15 halamanTally Prime Course Account and IntroAnkit Singh100% (1)
- Pitney BowesDokumen15 halamanPitney BowesPeter Sen Gupta100% (1)
- List of PlayStation Store TurboGrafx-16 GamesDokumen3 halamanList of PlayStation Store TurboGrafx-16 Gamesjavi83Belum ada peringkat
- Secospace USG2110 V100R001C03SPC200 User Guide (English Document)Dokumen10 halamanSecospace USG2110 V100R001C03SPC200 User Guide (English Document)Subarna GhimireBelum ada peringkat
- Sm-Sinamics S120Dokumen55 halamanSm-Sinamics S120preferredcustomerBelum ada peringkat
- Atoll 3.1.2 Exercises Radio-Libre PDFDokumen44 halamanAtoll 3.1.2 Exercises Radio-Libre PDFsaulernestofonsecaBelum ada peringkat
- Freebitco - in Script MULTIPLY BTC BONUS 2020Dokumen2 halamanFreebitco - in Script MULTIPLY BTC BONUS 2020Mohammadreza EmamiBelum ada peringkat
- Design and Development of Two Wheeler To Avoid Trible Riders and AccidentsDokumen6 halamanDesign and Development of Two Wheeler To Avoid Trible Riders and Accidentsvenkat krishnanBelum ada peringkat
- Hathway BillDokumen1 halamanHathway BillSanthoshkumar.Jayaram (CareerNet)Belum ada peringkat
- OM Chapter 3Dokumen28 halamanOM Chapter 3Tuấn TrườngBelum ada peringkat
- 03.10.12. Final Syllabus (M.SC Math)Dokumen24 halaman03.10.12. Final Syllabus (M.SC Math)Devil tigerBelum ada peringkat
- Oracle Cloud Migration & IntegrationDokumen30 halamanOracle Cloud Migration & IntegrationAqeel Nawaz100% (1)
- DM-HROD-2021-0340 Central Office Learning and Development Program COLDP Program Offerings For DepEd CO Personnel1Dokumen2 halamanDM-HROD-2021-0340 Central Office Learning and Development Program COLDP Program Offerings For DepEd CO Personnel1Nestor DawatonBelum ada peringkat
- CMT Eh15Dokumen50 halamanCMT Eh15Eletrônica Maia Comércio e ServiçosBelum ada peringkat
- The GD-04 "David" GSM Communicator: Table 1 - A Brief List of Programming SMS InstructionsDokumen8 halamanThe GD-04 "David" GSM Communicator: Table 1 - A Brief List of Programming SMS InstructionsJesús UrrietaBelum ada peringkat
- Cs HCM Person Basic Sync Hcm92 To Cs9Dokumen8 halamanCs HCM Person Basic Sync Hcm92 To Cs9cdahlinBelum ada peringkat