tmpEC5B TMP
Diunggah oleh
Frontiers0 penilaian0% menganggap dokumen ini bermanfaat (0 suara)
20 tayangan7 halamanAncient DNA helps in better understanding the evolution and migration pattern of humans throughout the world. Many technical difficulties arise during DNA extraction, PCR amplification and sequencing which make it difficult to work and interpret the analyses. Our preliminary result is encouraging, as we have successfully amplified 456 base pair PCR product of mitochondrial DNA hypervariable region I (HVR1) starting from position 15976 to 16431.
Deskripsi Asli:
Judul Asli
tmpEC5B.tmp
Hak Cipta
© © All Rights Reserved
Format Tersedia
PDF, TXT atau baca online dari Scribd
Bagikan dokumen Ini
Apakah menurut Anda dokumen ini bermanfaat?
Apakah konten ini tidak pantas?
Laporkan Dokumen IniAncient DNA helps in better understanding the evolution and migration pattern of humans throughout the world. Many technical difficulties arise during DNA extraction, PCR amplification and sequencing which make it difficult to work and interpret the analyses. Our preliminary result is encouraging, as we have successfully amplified 456 base pair PCR product of mitochondrial DNA hypervariable region I (HVR1) starting from position 15976 to 16431.
Hak Cipta:
© All Rights Reserved
Format Tersedia
Unduh sebagai PDF, TXT atau baca online dari Scribd
0 penilaian0% menganggap dokumen ini bermanfaat (0 suara)
20 tayangan7 halamantmpEC5B TMP
Diunggah oleh
FrontiersAncient DNA helps in better understanding the evolution and migration pattern of humans throughout the world. Many technical difficulties arise during DNA extraction, PCR amplification and sequencing which make it difficult to work and interpret the analyses. Our preliminary result is encouraging, as we have successfully amplified 456 base pair PCR product of mitochondrial DNA hypervariable region I (HVR1) starting from position 15976 to 16431.
Hak Cipta:
© All Rights Reserved
Format Tersedia
Unduh sebagai PDF, TXT atau baca online dari Scribd
Anda di halaman 1dari 7
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
E-ISSN: 0975-8232; P-ISSN: 2320-5148
IJPSR (2013), Vol. 4, Issue 9
(Research Article)
Received on 13 April, 2013; received in revised form, 03 June, 2013; accepted, 14 August, 2013; published 01 September, 2013
AMPLIFICATION AND SEQUENCING OF MITOCHONDRIAL DNA (HVR-I) EXTRACTED
FROM 1200 YEARS OLD HUMAN BONE SPECIMEN
Rajeev Kumar Pandey 1, 2, Deepankar Pratap Singh 3, Abid Ali 5, G Sudhakar 2 and V.R. Rao*1, 4
Anthropological survey of India 1, Mysore, Karnataka, India
Department of Human Genetics, Andhra University, Visakhapatnam 530003, Andhra Pradesh, India
S2-Functionnel Genomics, IBENS, UMR8197, Ecole Normale Superieure, 46 rue d'Ulm, 75005
Paris, France
Department of anthropology, University of Delhi, Delhi 110007, India
Department of Biotechnology, Bundelkhand University, Jhansi, Uttar Pradesh, India
Keywords:
Ancient DNA, PCR, Hypervariable
region I (HvrI), Contamination
Correspondence to Author:
Prof. V. R. Rao
Professor, Department of
Anthropology, University of Delhi,
North Campus, Delhi 110007, India
E -mail: drraovr2@gmail.com
ABSTRACT: Ancient DNA help in better understanding the
evolution and migration pattern of humans throughout the world. The
present work is a preliminary effort, to extract and amplify the ancient
DNA from human bone samples provided by the Anthropological
survey of India collected from the Himalayan region in 1952. Many
technical difficulties arise during DNA extraction, PCR amplification
and sequencing which make it difficult to work and interpret the
analyses. Our preliminary result is encouraging, as we have
successfully amplified 456 base pair PCR product of mitochondrial
DNA hypervariable region I (HVR1) starting from position 15976 to
16431. Our results confirm that the ancient bone specimens harbours
M haplogroup signature with considerable similarities to M21ab subgroup specific to East and Southeast Asia. Absence of negative
amplification confirms that our ancient mitochondrial DNA is
contamination free.
INTRODUCTION: An ancient DNA technology
provides new insight for anthropologist and
archaeologist to better understand the past
happenings and interpret it with the future
possibilities. The first ancient DNA study was done
in 1984, on museum specimen of the Quagga 1 but
the role of ancient DNA in humans comes into play
after the successful study on 2400-year old
Egyptian mummified material by Paabo and
colleagues in 1985 2.
QUICK RESPONSE CODE
DOI:
10.13040/IJPSR.0975-8232.4(9).3541-47
Article can be accessed online on:
www.ijpsr.com
Ancient DNA has generated new opportunities for
archaeological and anthropological investigators
like contemporary population history, their
migration patterns, attraction domestication of
plant, solving the historical mysteries and much 3, 4.
It is always a tough task to work with ancient
DNA, the two main hurdles which make it difficult
is degradation of DNA and contamination of
ancient DNA with contemporary DNA, the DNA
molecule present in the ancient remains may also
get destroyed during extraction due to physical and
chemical treatment. In 1989, the invention of the
polymerase chain reaction (PCR) 5 made it possible
to amplify ancient DNA and study even single
existing molecule which allows the number and
assortment of ancient DNA studies to expand
rapidly 6, 7, 8.
DOI link: http://dx.doi.org/10.13040/IJPSR.0975-8232.4(9).1000-06
International Journal of Pharmaceutical Sciences and Research
3541
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
E-ISSN: 0975-8232; P-ISSN: 2320-5148
This technique is extremely sensitive to amplify a
low copy number of DNA present in ancient
remains in a matter of hours 9, 10. Many researchers
are trying to develop more reliable techniques for
ancient human DNA analyses 11, 12, 13. The main
problem with the PCR amplification of ancient
DNA is presence of the inhibitors and damaged
DNA 14, 15. In order to overcome this problem
researchers have used Ampli Taq Gold TM
Polymerase to increase the efficiency of PCR
amplification which is highly expensive and low
cost Taq polymerase (Promega), high number of
PCR cycles, addition of BSA, etc are also used in
some studies . Ariffin et al (2006) 16 and Yang et al
(1998)17 were able to amplify mitochondrial DNA
from 400years old and 2000 years old human
skeletal remains and are the best examples of
amplifying the ancient DNA with high efficiency.
These samples were excavated from Himalayan
region, India by the Anthropological Survey of
India under the leadership of its Director, D.N.
Dutta-Mazumdar (1956) and carbon dating was
revealed that these samples are around 1200 years
old (Unpublished report 1959, Department of
anthropology, Kolkata) 18.
In the present study, the human bone samples were
excavated from the Himalayan regions by
Anthropological survey of India field team in the
1950s and carbon dating analysis and
morphological analyses showed that these samples
are 1200 years old and are human (Unpublished
report 1959, Department of anthropology, Kolkata)
18
. Since the excavated bone samples were placed
constantly at 4C after excavation, they seem to be
well preserved.
Contamination controls: All ancient DNA
extractions and PCR set ups should be conducted in
a dedicated laboratory that undergoes regular
decontamination 19. We have a dedicated Ancient
DNA laboratory where all rooms have ultraviolet
lamps fixed to ceilings for general room irradiation.
Glasses used in making doors and windows are
ultraviolet proof and fitted airtight. Clean filtered
air generated from an exclusive hepa-filter/AC unit
is flushed into the room finally with positive
pressure and restricted entry of people.
In this report, we follow a simple procedure for
mitochondrial DNA extraction and subsequent
PCR amplification using AmpliTaq Gold (Applied
Biosystems) polymerase followed by DNA
sequencing from 1200 year old human bone
sample. All ancient DNA research is carried out in
a dedicated laboratory that undergoes regular
decontamination process to remove the chance of
contamination of modern DNA or PCR products.
The DNA extractions were carried out on
powdered bone and were subjected to DNA
extraction by phenol-chloroform and ethanol
precipitated purification procedure followed by
PCR amplification.
MATERIALS AND METHODS
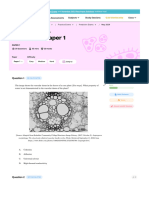
Ancient specimens: Ancient bone samples (Figure
1) were provided by the Anthropological Survey of
India and all experiments were performed at the
ancient DNA lab facility of the Centre for Cellular
and Molecular Biology (CCMB), Hyderabad.
FIGURE 1: ANCIENT BONE SPECIMENS CONSIDER
FOR
PRESENT
STUDY
PROVIDED
BY
ANTHROPOLOGICAL SURVEY OF INDIA
Personnel wear laboratory coats and face masks
and changes gloves regularly every time. All
general equipment and apparatus (e.g., centrifuges,
pipettes, gel electrophoresis) were dedicated for
this lab room for pre-PCR work (there is no shared
equipment). Protective clothing from a post-PCR
laboratory was never taken into the clean-room
facility. All solutions, PCR reagents, and primers
were kept in small carefully labelled aliquots
dedicated solely for work with one ancient DNA
collection.
Pre-PCR activities were spatially separated in the
laboratory and post PCR sequencing performed in a
different laboratory. Extraction of bone DNA was
performed using dedicated chamber that were
decontaminated by bleach followed by 70%
ethanol prior to the procedure so as to minimize
cross contamination with modern DNA as cited in
Kemp and Smith 2005 11.
International Journal of Pharmaceutical Sciences and Research
3542
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
Extraction of ancient bone DNA: The bone
surface was shaved by sterile scalpel blade to
remove dirt, soil, other foreign material and then
wiped with alcohol. The bone samples were then
exposed under UV lamp in the aspiration hood for
2hrs. About 4 mm square of cortical bone surface
was removed with a sterile scalpel from all
samples, followed by immersion of fragmented
bones in 10% bleach solution for 10 min and
washing with 70% alcohol 11. The cleaned bone
fragments were mechanically pulverized into a fine
powder in sterile pestle-mortar.
Approximately 1 milligram (mg) of powdered bone
was taken for DNA extraction. In this study, due to
the importance of the respective samples, we were
allowed to extract the mtDNA from a total of 1
grams of powdered bone. The powdered bone was
UV irradiated prior to DNA extraction. The
samples (500mg) were then soaked in 5ml of 0.5M
EDTA (pH 8.0) along with 200l of 20mg/ml
Proteinase K and 120l of 10% SLS (Sodium
Lauryl Sulphate) followed by incubation at 55C
for overnight (24 hours).
Two different methods were employed for the
extraction of DNA from the powdered samples. In
the first method decalcified samples were
processed with Banglore genei kit as follows
Extraction using Bangalore Genei Kit: After
overnight incubation, pellet was washed twice with
sterile water and centrifuged for 15min at 4000
rpm. After discarding the supernatant, lysis buffer 1
and proteinas K were added as mentioned in
Bangalore geni manual then incubated at 50C for
24-48 hours. On the third day lysis buffer was
added and centrifuged at 5000 rpm for 10min.
Supernatant was collected into fresh tube in
addition with binding buffer and passed it through
the column provided in kit by centrifuging at
10,000rpm for 1min. Column was washed by wash
buffer I followed by wash buffer II . Finally the
DNA is eluted from the column by adding 40 l of
elution buffer.
The extraction was also done with Phenolchloroform method.
Extraction using Phenol-chloroform method: In
this method decalcified samples were lysed in 2ml
of Lysis buffer (10mM tris HCl ph 8.0, 1mM NaCl)
E-ISSN: 0975-8232; P-ISSN: 2320-5148
at 60C for 24 hrs, which was later subjected to
phenol-chloroform, chloroform-isoamyl alcohol
extraction (Kalmar et al., 2000 with modifications)
20
. At last, the aqueous phase was concentrated by
centrifugation driven dialysis using Centricon-30
micro concentrators from Amicon.
The concentrate, which was about 0.7ml, purified
by Gene Clean Spin Kit and later eluted in 0.1m
Tris-EDTA. Approximately 40l of DNA extract
was obtained.
Amplification of ancient bone mtDNA: The
overall success of the extraction procedure was
assessed based on the ability to amplify a long
stretch of 456 base pairs target sequence at
Hypervariable region I of mtDNA which is not
usual case with ancient DNA samples 20. This
could be possible because of proper purification of
DNA, which removed prominent PCR inhibitors
efficiently. The following primers were designed
and synthesized (Sigma Aldrich Chemicals Pvt.
Ltd, Bangalore, India) for PCR amplification:
F15976 5CTCCACCATTAGCACCCAAAGC3
as
the
forward
primer
and
R16431
5GCGGGATATTGATTTCACGG 3 as the
reverse primer.
PCR amplification was carried out using the
Applied biosystem Thermocycler Model 2720
(ABI) in a 20 l reaction volume containing 3L
of re-precipitated template, 100M each of dNTPs,
4 pM of each primer, 1X PCR buffer [100mM TrisHCL, pH 8.3 (at 250C), 500mM KCl, 15 mM
MgCl2, 0.01%(w/v) gelatin, bovine serum albumin
(BSA, 10 mg/ml, New England Bio Labs) and 0.5
units of AmpliTaq Gold (Applied Biosystems). The
PCR reaction was carried out in an ABI_2720
under the following conditions: 96C for 10
minutes, 35 cycles at 96C for 45 seconds, 57C for
1 minute, and 72C for 2 minutes, and a final
extension at 72C for 20 minutes.
Finally 10 l PCR products of ancient DNA were
electrophoresed at 120V in 2% agarose gel.
Agarose gel was prepared in 1X Tris-acetateEDTA (TAE) buffer containing ethidium bromide
(EtBr). After 30 Min. of run halfway the gel was
observed under ultraviolet light of UV transilluminator and photographed (Figure 2).
International Journal of Pharmaceutical Sciences and Research
3543
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
E-ISSN: 0975-8232; P-ISSN: 2320-5148
The variations detected in the Ancient DNA were
checked in the mitochondrial databases such as
mitochondria
(http://www.mitochondria.org/
mitologial.php) for their signicance.
FIGURE 2: AGAROSE DNA ELECTROPHORESIS
(2%) OF PCR REACTION PRODUCTS [Lane1: phage
DNA marker (100bp), Lane 2: contain Ancient DNA
extracted by Phenol- Chloroform method (single faint band),
Lane 3: contain Ancient DNA extracted by Bangalore Genei
Kit (no amplification), Lane 4: Positive Control, Lane 5:
Negative control, and Lane 6: Blank]
ExoSAP treatment: In order to sequence, the
amplicons were treated with exonuclease-1 and
Shrimp Alkaline Phosphatase (ExoSAP-IT; USB
Corporation, Cleveland, Ohio, USA). 5l of PCR
Product is properly mixed with 2l ExoSAP-IT and
incubate (Thermocycler machine) at 37C for
15min (Activation of enzyme) followed by 80C
for 30min (Inactivation of enzyme) to remove the
extra nucleotides and primers (dimer) without loss
of amplified PCR product.
Sequencing of ancient bone mitochondrial DNA
(HVR-I): ExoSAP treated PCR products were
directly sequenced twice using the ABI Prism
3700 DNA analyzer (Applied Biosystems).The
sequencing PCR was carried out in GeneAmp 9600
thermocycler (Perkin-Elmer) for 3hours using the
BigDye Terminator v3.1 ready reaction kit
(Applied Biosystems, Foster City, CA, USA) and
analyzed in an ABI 3730xl automated DNA
Analyzer (Applied Biosystems). Sequences of
ancient DNA were carefully edited and aligned
with revised Cambridge reference sequence (rCRS)
and compared with the mitochondrial HVR-I
region using sequence analysis ,AutoAssembler
tool-version 2.1 (Perkin-Elmer, Foster City,CA,
USA) and Codon Code Aligner version 2.0.5
(Codon Code Corporation, Dedham, MA, USA)
(Figure 3).
FIGURE 3: THE SEQUENCE ELECTROPHEROGRAM
OF mtDNA POLYMORPHISMS OBSERVED. A. The
wild type sequence showing (arrow) A at the nucleotide
position 16037. B. Sequence of an ancient DNA showing
polymorphic allele Cat the nucleotide position 16184. C.
Sequence of an ancient DNA showing polymorphic allele
Cat the nucleotide position 16223.
RESULTS: Primers were specially designed to
amplify mitochondrial DNA hypervariable region I
on position 15976 to 16431 for 456 base pairs PCR
product. A band approximately 456 base pairs was
amplified in the samples containing ancient bone
International Journal of Pharmaceutical Sciences and Research
3544
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
(Figure 2, Lane 2) and positive control (Figure 2,
Lane 4) whereas negative and blank controls were
unable to amplify the 456 base pairs band (Figure
2, Lane 5, 6). However, Ancient DNA and positive
control (+ve) samples showed the presence of
primer-dimers (Figure 2, Lanes 2-4) during PCR
amplification as expected due to the sequences of
the primers 30.
In Figure 2 Lane 2, After 35 amplification cycles a
single weak band of 456 bp provides an evidence
of a successful amplification of DNA from an
ancient bone. PCR products from successful
amplifications were subjected to DNA sequencing
after exosap treatment. The sequence electropherogram of mtDNA shows polymorphisms at the
nucleotide position A16037G (Figure-3, A),
C16184T (Figure-3, B), C16223T (Figure-3, C) in
ancient DNA. BLAST analysis confirms that
ancient DNA was 99% identical to the human
mitochondrial
displacement
loop
(D-loop)
sequence.
The phylogenetic analysis revealed haplogroup M
to be the major haplogroup and considerable
similarities with M21ab sub-group from region of
India, China, Pakistan, Japan population for our
ancient
DNA
sample.
The
haplogroup
nomenclature was based on published information
21
. The samples were not showing any
heteroplasmy which also confirms that our ancient
mitochondrial DNA is not contaminated.
Control amplification: The present study included
control amplification in each step to assess any
possible
contamination.
Negative
control
amplification, to check the purity of the PCR
reagents with no DNA added (Figure 2, Lane6).
Blank control amplification is used to check the
purity of the extraction reagents with no bone
added followed by PCR amplifications (Figure 2,
Lane5) 15, 17, 22, 23.
Positive control amplification used to monitor the
success of Thermocycler reactions and PCR
components (Figure 2, Lane4).
DISCUSSION: Working with ancient DNA is very
expensive, time-consuming, needs more amount of
sample (approximately 5g of bone powder) and
destructive undertaking. Every time the results
were distrustful to be a contaminant free.
E-ISSN: 0975-8232; P-ISSN: 2320-5148
The reliability of ancient DNA is always
questionable due to several reasons. Furthermore,
the ancient DNA loses its integrity and decomposes
with an irreversible loss of nucleotide sequence
information relative to the modern DNA. From
available studies it was shown that Svante Paabo
(Nature news) 24 on 38,000 to 44,000 years old
bones samples and Rasmussen on 4,000-year-old
hair Palaeo-Eskimo samples were able to yield
DNA that could be enzymatically amplified and
sequenced 25.
In our earlier study we are able to increase the
efficiency of PCR amplification from old bone
specimens by optimizing PCR component as well
as PCR condition for amplify HVR1 region26.We
choose slightly lower number of PCR cycles to
avoid the unspecific amplification with use of
standard DNA polymerase i.e AmpliTaq Gold
polymerase (studies by other groups showed that a
strong polymerase to amplify the ancient DNA)
and addition of BSA. In this present study, absence
of amplification in negative control confirmed that
there is no modern DNA contamination during
extraction and PCR procedures.
Although negative control shows primer-dimer but
does not proves modern DNA contamination. From
the gel picture, it was clear that amplified product
similar in size to ancient D-loop mitochondrial
hypervariable region I. This indicates that the
procedure used
for DNA extraction is able to
produce a pure DNA from ancient bones that can
be amplified and sequenced. Contamination of
ancient samples by modern DNA molecules is a
serious problem. Human remains are particularly
difficult to work with; it gets easily contaminated at
any step of excavation, extraction and finally PCR
setup 10, 17.
For working with ancient DNA we have to follow
the strict rule to avoid contamination. In many
studies it was shown that researcher have the
dedicated laboratory for ancient DNA extraction
but they use the reagents, buffers, tubes and
chemicals that take place outside of the laboratory
can be contaminated by modern DNA 22. In this
study, we have strictly followed all decontaminated
procedures to minimize or eliminate modern DNA
contamination in the sample. There is no
amplification product found in the negative control
(Figure 2, Lane 6) and also sample processed with
International Journal of Pharmaceutical Sciences and Research
3545
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
Bangalore Genei kit not showed any amplification
(Figure 2, Lane3). For our study, we have used the
dedicated laboratory as well as laminar chamber to
minimize the contamination. In addition, we have
prepared all reagents in this dedicated laboratory
only and also consumable items and equipments
needed for ancient DNA work were double
autoclaved and exposed
to UV irradiation.
Furthermore, Fumigation and disinfectants were
regularly done in the lab to eliminate any surface
contamination. In this study, we also performed
different extraction method with some modification
based on different reports. The good preservation
of the samples and contamination free protocols
have been key factors for the success of this study.
The analysis of mitochondrial DNA (mtDNA) has
been proven to be a powerful tool in the
understanding of human evolution and a study by
Hagelberg had confirmed that this is a potent tool
for identification of variations in mitochondrial
DNA from human remains for chronological
investigations. The main reason for recovering
mitochondrial DNA (mtDNA) from ancient Human
bone specimen is its lack of recombination,
contains high copy number, shows maternal mode
of inheritance and help to calculate the divergence
time elapsed. Finally small size and simple genome
organization make it easier to study. The high copy
number of mitochondria per cell increases the
probability to amplify the ancient DNA. There are
enough
evidences
that
show
successful
mitochondrial DNA analyses on very old human
remains 4, 27, 28. In our study, we have chosen the
mitochondrial Hypervariable segments, HVRI
(positions 15976 - 16431) region as targets for PCR
amplification (Figure 2, Lane 2). Best combination
of DNA polymerase (Ampligold Taq) & other PCR
components (BSA) provides sufficient amount of
PCR product for DNA sequencing. For our smaller
quantities of ancient DNA extracted from bone
specimen, we used higher number of cycles to
produce high number of copies 15, 17, 22. Almost all
studies of human evolution based on mtDNA
sequencing have been confined to the control
region also called the D-loop or the displacement
loop. HVRI and HVRII data can provide useful
insights about inter and intra-specific population
variation. Displacement loop (D-loop) is noncoding regions of mitochondrial DNA which
constitutes about 7% of the mitochondrial genome.
The sequence and size of HVRI (D-loop) varies
E-ISSN: 0975-8232; P-ISSN: 2320-5148
from species to species29. So on this basis expected
sizes of ancient DNA PCR product showed that the
DNA originated from human. The sequence
generated by the amplified ancient DNA showed
99% identical with the hypervariable region I of Dloop region of the mitochondria with the absence of
recombination. Furthermore, sequences were
analysed using NCBI databases showed that
ancient DNA was from human DNA only.
In summary, we managed to extract mitochondrial
DNA from approximately 1200 year old bone
specimen recovered by the Anthropological survey
of India from the Himalayan region. The carbon
dating conducted as reported showed that these
samples are 1200 years old (unpublished data).
Since, we were able to extract enough amount of
DNA from these samples which is sufficient for
amplifying 4-5 PCR reactions, the DNA sequences
gave 99% homology with human mitochondrial
HVR-I DNA.
Acknowledgment
The authors thank Dr K.Thangaraj, Scientist,
Centre for Cellular and Molecular Biology, Uppal
Road, Hyderabad, India for providing all the lab
facilities. The financial assistance to Mr. Rajeev
Kumar Pandey by Anthropological survey of India
(AnSI) as JRF and Council of Scientific &
Industrial Research (CSIR) as SRF is gratefully
acknowledged.
References:-
International Journal of Pharmaceutical Sciences and Research
1.
2.
3.
4.
5.
6.
7.
8.
Higuchi R, Bowman B, Freiberger M, Ryder OA, and
Wilson AC: DNA sequences from the quagga, an extinct
member of the horse family. Nature 1984; 312 (5991):
2824.
Paabo S: Molecular cloning of ancient Egyptian mummy
DNA. Nature 1985; 314:644-45.
Brown TA, Allaby RG, Sallares R, and Jones G: Ancient
DNA in charred wheats: taxonomics identification of
mixed and single grains. Ancient Biomolecules 1998; 2:
185-193.
Hagelberg E, Sykes B, and Hedges R: Ancient bone
amplified. Nature 1989; 342: 485.
Mullis KB, Faloona, F: Specific synthesis of DNA in vitro
via a polymerase catalysed chain reaction. Methods
Enzymol 1987; 155:335-350.
Paabo S: Ancient DNA; extraction, characterization,
molecular cloning and enzymatic amplification. Proc. Natl
Acad. Sci. USA 1989; 86:19391943.
Paabo S, Wilson AC: Polymerase chain reaction reveals
cloning artefacts. Nature 1988; 334:387388.
Thomas R H, Schaffner W, Wilson A C, Paabo S: DNA
phylogeny of the extinct marsupial wolf. Nature 1989;
340:465467.
3546
Pandey et al., IJPSR, 2013; Vol. 4(9): 3541-3547.
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
19.
20.
Innis MA, Gelfand DH, Sninsky JJ: In A guide to methods
and amplifications in PCR protocols. San Diego CA
Academic Press 1990; 3-12.
Yang DY, Watt K: Contamination controls when preparing
archaeological remains for ancient DNA analysis. Journal
of Archaeological Science 2005; 32: 331-336.
Kemp BM, Smith DG: Use of bleach to eliminate
contaminating DNA from the surface of bones and teeth.
Forensic Science International 2005; 154: 53-61.
Sampietro ML, Gilbert MT, Lao O, Caramelli D, Lari M,
Bertranpetit J, Lalueza- Fox C: Tracking down human
contamination in ancient human teeth. Mol Biol Evol
2006; 23:1801-1807
Helgason A, Plsson S, Lalueza-Fox C, Ghosh S,
Sigurethardottir S, Baker A, Hrafnkelsson B, Arnadottir L,
Thornorsteinsdottir U, Stefansson K.: A Statistical
Approach to Identify Ancient Template DNA. J Mol Evol
2007; 65:92-102.
Herrmann R G, Hummel S. Ancient DNA: Recovery and
Analysis of Genetic Material from Paleontological,
Archaeological, Museum, Medical and Forensic
Specimens. New York, NY: Springer Verlag 1994.
Yang DY, Eng B, Saunders SR: Hypersensitive PCR,
ancient human mtDNA and contamination. Human
Biology 2003; 75: 355-364.
Ariffin S H Z, Wahab RM A, Zamrod Z, Sahar S, Razak
M F A , Ariffin E J,Senafi S: Molecular Archaeology of
Ancient Bone From 400 Year Old Shipwreck. AsPac J.
Mol. Biol. Biotechnol 2007; 15 (1):27-31.
Yang DY, Eng B, Waye JS, Dudar JC, and Saunders SR:
Technical note: Improved DNA extraction from ancient
bones using silica-based columns: American Journal of
Physical Anthropology 1998; 105: 539-543.
Unpublished report: Anthropological Investigation in
Rupkund. Anthropologcial survey of India Library.
Department of anthropology, government of India, India
museum Calcutta 1959; 17434: 1-50.
Kemp BM, Smith DG: Ancient DNA Methodology:
Thoughts from Brian M. Kemp and David Glenn Smith on
Mitochondrial DNA of Protohistoric Remains of an
Arikara Population from South Dakota. Human Biology
2010; 82(2): 227-238.
Kalmar T, Bachrati C, Marcsik A, Rask I: A simple and
efficient method for PCR amplifiable DNA extraction
from ancient bones. Nucleic Acids Res 2000; 28(12): e67.
E-ISSN: 0975-8232; P-ISSN: 2320-5148
21. Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh
VK, Rasalkar AA, Singh L: Reconstructing the origin of
Andaman Islanders. Science 2005a; 308: 996.
22. Eshleman J, Smith DG: Use of DNase to eliminate
contamination in ancient DNA analysis. Electrophoresis
2001; 22: 4316-4319.
23. Kalmar T, Bachrati CZ, Marcsik A, Rasko I: A simple and
efficient method for PCR amplifiable DNA extraction
from ancient bones. Nucleic Acids Res 2000; 28: e67.
24. Nature news: Ancient DNA set to rewrite human history.
Nature 2010; 465:148-149.
25. Rasmussen M, Li Y, Lindgreen S, Pedersen J S,
Albrechtsen A, Moltke I, Metspalu M, Metspalu E,
Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert M T,
Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson
AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED,
Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen
M, Orlando L, Kristiansen K, Bak M, Tommerup N,
Bendixen C, Pierre TL, Grnnow B, Meldgaard M,
Andreasen C, Fedorova SA, Osipova LP, Higham TF,
Ramsey CB, Hansen TV, Nielsen FC, Crawford MH,
Brunak S, Sicheritz-Pontn T, Villems R, Nielsen R,
Krogh A, Wang J, Willerslev E: Ancient human genome
sequence of an extinct Palaeo-Eskimo. Nature 2010;
463(7282):757-62.
26. Pandey RK, Singh DP, Sudhakar G, Thangaraj K, Rao
VR: Standardization of PCR conditions for an Ancient
DNA Amplification IJHS 2012; 9(1):102-109
27. Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking
M, Paabo S: Neanderthal DNA sequences and the origin of
modern human. Cell 1997; 90: 19-30.
28. Ovchinnikov IV, Gotherstrom A, Romanova VM ,
Kharitonov VM, Liden K, Goodwin W: Molecular
analysis of Neanderthal DNA from the northern Caucasus.
Nature 2000; 404: 490-493.
29. Fernandez-Silva P, Enriquez JA, Montoya J: Replication
and transcription of mammalian mitochondrial DNA. Exp.
Physiol 2003; 881: 41-56.
30. Tibor K, Csand Z, Bachrati AM, Istvn R: A simple and
efficient method for PCR amplifiable DNA extraction
from ancient bones. Nucleic Acids Research 2000; 28:
E67-E67.
How to cite this article:
Pandey RK, Singh DP, Ali A, Sudhakar G and Rao VR: Amplification and Sequencing of Mitochondrial DNA (HVR-I)
extracted from 1200 years old human bone specimen. Int J Pharm Sci Res 2013: 4(9); 3541-3547. doi: 10.13040/IJPSR.
0975-8232.4(9).3541-47
All 2013 are reserved by International Journal of Pharmaceutical Sciences and Research. This Journal licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
This article can be downloaded to ANDROID OS based mobile. Scan QR Code using Code/Bar Scanner from your mobile.
(Scanners are available on Google Playstore)
International Journal of Pharmaceutical Sciences and Research
3547
Anda mungkin juga menyukai
- tmp3CAB TMPDokumen16 halamantmp3CAB TMPFrontiersBelum ada peringkat
- tmpCE8C TMPDokumen19 halamantmpCE8C TMPFrontiersBelum ada peringkat
- tmpEFCC TMPDokumen6 halamantmpEFCC TMPFrontiersBelum ada peringkat
- tmpF178 TMPDokumen15 halamantmpF178 TMPFrontiersBelum ada peringkat
- tmp72FE TMPDokumen8 halamantmp72FE TMPFrontiersBelum ada peringkat
- Tmpa077 TMPDokumen15 halamanTmpa077 TMPFrontiersBelum ada peringkat
- tmpE7E9 TMPDokumen14 halamantmpE7E9 TMPFrontiersBelum ada peringkat
- tmp80F6 TMPDokumen24 halamantmp80F6 TMPFrontiersBelum ada peringkat
- tmp6F0E TMPDokumen12 halamantmp6F0E TMPFrontiersBelum ada peringkat
- Tmp1a96 TMPDokumen80 halamanTmp1a96 TMPFrontiersBelum ada peringkat
- tmpE3C0 TMPDokumen17 halamantmpE3C0 TMPFrontiersBelum ada peringkat
- tmp6382 TMPDokumen8 halamantmp6382 TMPFrontiersBelum ada peringkat
- tmpFFE0 TMPDokumen6 halamantmpFFE0 TMPFrontiersBelum ada peringkat
- tmpF407 TMPDokumen17 halamantmpF407 TMPFrontiersBelum ada peringkat
- tmpF3B5 TMPDokumen15 halamantmpF3B5 TMPFrontiersBelum ada peringkat
- tmpC0A TMPDokumen9 halamantmpC0A TMPFrontiersBelum ada peringkat
- tmp998 TMPDokumen9 halamantmp998 TMPFrontiersBelum ada peringkat
- tmp60EF TMPDokumen20 halamantmp60EF TMPFrontiersBelum ada peringkat
- tmp8B94 TMPDokumen9 halamantmp8B94 TMPFrontiersBelum ada peringkat
- tmpA0D TMPDokumen9 halamantmpA0D TMPFrontiersBelum ada peringkat
- tmpD1FE TMPDokumen6 halamantmpD1FE TMPFrontiersBelum ada peringkat
- tmp4B57 TMPDokumen9 halamantmp4B57 TMPFrontiersBelum ada peringkat
- tmp9D75 TMPDokumen9 halamantmp9D75 TMPFrontiersBelum ada peringkat
- tmp37B8 TMPDokumen9 halamantmp37B8 TMPFrontiersBelum ada peringkat
- tmp27C1 TMPDokumen5 halamantmp27C1 TMPFrontiersBelum ada peringkat
- tmpC30A TMPDokumen10 halamantmpC30A TMPFrontiersBelum ada peringkat
- tmpB1BE TMPDokumen9 halamantmpB1BE TMPFrontiersBelum ada peringkat
- Tmp75a7 TMPDokumen8 halamanTmp75a7 TMPFrontiersBelum ada peringkat
- tmp3656 TMPDokumen14 halamantmp3656 TMPFrontiersBelum ada peringkat
- tmp2F3F TMPDokumen10 halamantmp2F3F TMPFrontiersBelum ada peringkat
- Shoe Dog: A Memoir by the Creator of NikeDari EverandShoe Dog: A Memoir by the Creator of NikePenilaian: 4.5 dari 5 bintang4.5/5 (537)
- Never Split the Difference: Negotiating As If Your Life Depended On ItDari EverandNever Split the Difference: Negotiating As If Your Life Depended On ItPenilaian: 4.5 dari 5 bintang4.5/5 (838)
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureDari EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FuturePenilaian: 4.5 dari 5 bintang4.5/5 (474)
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeDari EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifePenilaian: 4 dari 5 bintang4/5 (5782)
- Grit: The Power of Passion and PerseveranceDari EverandGrit: The Power of Passion and PerseverancePenilaian: 4 dari 5 bintang4/5 (587)
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceDari EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RacePenilaian: 4 dari 5 bintang4/5 (890)
- The Yellow House: A Memoir (2019 National Book Award Winner)Dari EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Penilaian: 4 dari 5 bintang4/5 (98)
- On Fire: The (Burning) Case for a Green New DealDari EverandOn Fire: The (Burning) Case for a Green New DealPenilaian: 4 dari 5 bintang4/5 (72)
- The Little Book of Hygge: Danish Secrets to Happy LivingDari EverandThe Little Book of Hygge: Danish Secrets to Happy LivingPenilaian: 3.5 dari 5 bintang3.5/5 (399)
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryDari EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryPenilaian: 3.5 dari 5 bintang3.5/5 (231)
- Team of Rivals: The Political Genius of Abraham LincolnDari EverandTeam of Rivals: The Political Genius of Abraham LincolnPenilaian: 4.5 dari 5 bintang4.5/5 (234)
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaDari EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaPenilaian: 4.5 dari 5 bintang4.5/5 (265)
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersDari EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersPenilaian: 4.5 dari 5 bintang4.5/5 (344)
- The Emperor of All Maladies: A Biography of CancerDari EverandThe Emperor of All Maladies: A Biography of CancerPenilaian: 4.5 dari 5 bintang4.5/5 (271)
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyDari EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyPenilaian: 3.5 dari 5 bintang3.5/5 (2219)
- The Unwinding: An Inner History of the New AmericaDari EverandThe Unwinding: An Inner History of the New AmericaPenilaian: 4 dari 5 bintang4/5 (45)
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreDari EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You ArePenilaian: 4 dari 5 bintang4/5 (1090)
- Rise of ISIS: A Threat We Can't IgnoreDari EverandRise of ISIS: A Threat We Can't IgnorePenilaian: 3.5 dari 5 bintang3.5/5 (137)
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)Dari EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Penilaian: 4.5 dari 5 bintang4.5/5 (119)
- Her Body and Other Parties: StoriesDari EverandHer Body and Other Parties: StoriesPenilaian: 4 dari 5 bintang4/5 (821)
- Prepared and Presented By: Fatuma Felix (B.pharm, M.SC.) China Pharmaceutical University 15-06-2016Dokumen24 halamanPrepared and Presented By: Fatuma Felix (B.pharm, M.SC.) China Pharmaceutical University 15-06-2016Fatma Felix MayugiBelum ada peringkat
- Potential Targets For Antifungal Drug Discovery - Creative Biolabs - 1619624328334Dokumen6 halamanPotential Targets For Antifungal Drug Discovery - Creative Biolabs - 1619624328334DicksonBelum ada peringkat
- UCLA DEV DEPT Health - SciencesDokumen1 halamanUCLA DEV DEPT Health - SciencesCapital_and_MainBelum ada peringkat
- HERU SETIAWAN-Week-4 Assigment Gene InteractionDokumen2 halamanHERU SETIAWAN-Week-4 Assigment Gene InteractionHeru SetiawanBelum ada peringkat
- Updatedsignature Assignment Bio1610Dokumen4 halamanUpdatedsignature Assignment Bio1610api-355514096Belum ada peringkat
- KMIslam ELISADokumen9 halamanKMIslam ELISADlstmxkakwldrl VousmevoyezBelum ada peringkat
- A KaryotypeDokumen2 halamanA KaryotypeHOLA AHORABelum ada peringkat
- Superscript IV Vilo Optimal RT QPCR White Paper PDFDokumen8 halamanSuperscript IV Vilo Optimal RT QPCR White Paper PDFGabyMezaCarrascoBelum ada peringkat
- Understanding Cell Structure and Function Through Biology QuestionsDokumen6 halamanUnderstanding Cell Structure and Function Through Biology QuestionsazwaBelum ada peringkat
- CH 25 Basic Genetics: Chapter TestDokumen11 halamanCH 25 Basic Genetics: Chapter Testedmond 黃Belum ada peringkat
- Lecture 5 PharmacovigilanceDokumen32 halamanLecture 5 Pharmacovigilancephoto copyhemnBelum ada peringkat
- Sbstta 18 Inf 03 enDokumen63 halamanSbstta 18 Inf 03 enPasta LoverBelum ada peringkat
- CTD 05-2008 en PDFDokumen303 halamanCTD 05-2008 en PDFMarija MarinkovićBelum ada peringkat
- IB Biology SL - 2024 Prediction Exam - May 2024 Paper 1Dokumen16 halamanIB Biology SL - 2024 Prediction Exam - May 2024 Paper 1Christy HuynhBelum ada peringkat
- Nesters Microbiology A Human Perspective 8th Edition Anderson Solutions ManualDokumen2 halamanNesters Microbiology A Human Perspective 8th Edition Anderson Solutions Manualcoactiongaleaiyan100% (20)
- Bioinformatics History of BioinformaticsDokumen10 halamanBioinformatics History of BioinformaticsSir RutherfordBelum ada peringkat
- Eukaryotic TranscriptionDokumen34 halamanEukaryotic TranscriptionKanaka lata SorenBelum ada peringkat
- Amyloid Vaccine For Alzheimer's Disease - Is It Feasible by Supreet Khare, Deeksha Seth, Shrayash KhareDokumen4 halamanAmyloid Vaccine For Alzheimer's Disease - Is It Feasible by Supreet Khare, Deeksha Seth, Shrayash Khareijr_journalBelum ada peringkat
- Lesson Plan in Science 10: Junior High School DepartmentDokumen4 halamanLesson Plan in Science 10: Junior High School DepartmentJoanne GodezanoBelum ada peringkat
- ProlpDokumen25 halamanProlpherryBelum ada peringkat
- Clinical Bacteriology: Fawad Mahmood M.Phil. Medical Laboratory SciencesDokumen8 halamanClinical Bacteriology: Fawad Mahmood M.Phil. Medical Laboratory SciencesFawad SawabiBelum ada peringkat
- Biochemistry 7th Edition Campbell Test Bank DownloadDokumen33 halamanBiochemistry 7th Edition Campbell Test Bank Downloaddaddockstudderyxeq100% (32)
- Learning Log L-L Matrix: Julyvern C. SimyunnDokumen7 halamanLearning Log L-L Matrix: Julyvern C. SimyunnMary Cecile H. PlatonBelum ada peringkat
- PRE-BOARD EXAMINATION, January 2015 Class XII Biology (044) : G.G.N. Public School, Rose Garden, LudhianaDokumen5 halamanPRE-BOARD EXAMINATION, January 2015 Class XII Biology (044) : G.G.N. Public School, Rose Garden, LudhianaSamita BhallaBelum ada peringkat
- Molecular Basis of Inheritance: By: Dr. Anand ManiDokumen130 halamanMolecular Basis of Inheritance: By: Dr. Anand ManiIndu Yadav100% (3)
- Dna FingerprintingDokumen131 halamanDna FingerprintingArnen PasaribuBelum ada peringkat
- Biochemistry - Module 1Dokumen6 halamanBiochemistry - Module 1ricky fecaraBelum ada peringkat
- Chemical Biology: Semester - Iii and Vii 2017-18Dokumen12 halamanChemical Biology: Semester - Iii and Vii 2017-18Yogesh ShekhawatBelum ada peringkat
- CloningDokumen6 halamanCloningkikkabuttigieg1466Belum ada peringkat
- Cancer JournalDokumen13 halamanCancer Journalhasna muhadzibBelum ada peringkat