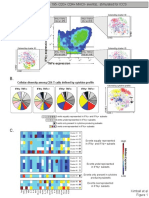
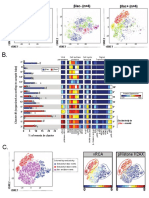
Kimball Et Al 2019, Figure 2
Diunggah oleh
Abby KimballJudul Asli
Hak Cipta
Format Tersedia
Bagikan dokumen Ini
Apakah menurut Anda dokumen ini bermanfaat?
Apakah konten ini tidak pantas?
Laporkan Dokumen IniHak Cipta:
Format Tersedia
Kimball Et Al 2019, Figure 2
Diunggah oleh
Abby KimballHak Cipta:
Format Tersedia
A.
Region Selection using PhenochartTM
1. Select and designate ROIs 2. Define ROI coordinates 3. Scan ROIs at higher
magnification, export
Select ROIs utilizing: image as .im3 file
#1 1) computer assisted or
#2 2) user defined methods
X: 50.92 mm, Y: 6.00 mm
#3 Hovering the cursor in
#4 ...[50961, 6064].im3
the middle of the ROI will
provide approximate coordinates
File name contains exact
Number ROIs according to position PhenochartTM coordinates
B. Spectral Unmixing & Visualization using inForm®
1. Build spectral library 2. Spectrally unmix .im3 files 3. Examine signal fluorescence
across all channels
Opal 520 Opals 540-690
DAPI
X
CD3 CD4 B220
FoxP3 CD8 F4/80
Autofluorescence Signal No signal
ex: 520 single stain
CD3 Examine signal across
FoxP3
CD4
CD8
Assign IDs and 4. Confirm signals make biological sense all channels to identify
B220 pseudocolors spectral bleedover
F4/80
DAPI for each stain
Autofluorescence Unexpected colocalization
of markers suggests
Utilize appropriate single
staining artifacts and/or
stain, unstained, and DAPI
issues with panel
controls for spectral unmixing F4/80 CD3
design/staining protocol
C. Cell Segmentation using inForm®
1. Empirically define settings to
Default Settings
identify cellular compartments
(i.e. nucleus, membrane)
Change settings while Unsegmented Image Fill holes Fill holes Fill holes Fill holes Fill holes
visualizing the results Roundness Roundness
Min-Max size
Roundness Roundness
Min-Max size
Roundness
Min-Max size Min-Max size Min-Max size
of segmentation Refine splitting Refine splitting Refine splitting Refine splitting Refine splitting
2. Apply segmentation settings to all images
Nucleus
Membrane
Green = predicted nucleus
Red = predicted membrane
3. Examine segmentation results on all ROIs
Retrain iteratively as needed
Anda mungkin juga menyukai
- Gender ClassificationDokumen5 halamanGender ClassificationAdedayo tunjiBelum ada peringkat
- Andor Istar 334 SpecificationsDokumen7 halamanAndor Istar 334 Specifications马马淼Belum ada peringkat
- Ijet V3i4p19Dokumen6 halamanIjet V3i4p19International Journal of Engineering and TechniquesBelum ada peringkat
- EC8002 Multimedia Compression and Communication Notes 2Dokumen109 halamanEC8002 Multimedia Compression and Communication Notes 2arivasanthBelum ada peringkat
- Mohini Dey - CapstoneDokumen52 halamanMohini Dey - CapstoneGautham Krishna KongattilBelum ada peringkat
- Journalpaper PDFDokumen8 halamanJournalpaper PDFkarpagaabiramiBelum ada peringkat
- UQ3 Techdata en 201905 PDFDokumen13 halamanUQ3 Techdata en 201905 PDFVishnuMaliBelum ada peringkat
- Potree ENDokumen16 halamanPotree ENjvaloosBelum ada peringkat
- Audio Fingerprinting Based On MultipleDokumen4 halamanAudio Fingerprinting Based On Multiplekvkumar128Belum ada peringkat
- Evaluating Spatial Sound SystemsDokumen32 halamanEvaluating Spatial Sound SystemsMark BockoBelum ada peringkat
- SPWLA - ML Workshop CheatsheetDokumen2 halamanSPWLA - ML Workshop CheatsheetSaeful Ghofar Zamianie PutraBelum ada peringkat
- Speaker Recognition: Tony (Mr. T)Dokumen10 halamanSpeaker Recognition: Tony (Mr. T)Tran TrungBelum ada peringkat
- Gender Detection by Voice Using Deep LearningDokumen5 halamanGender Detection by Voice Using Deep LearningInternational Journal of Innovative Science and Research TechnologyBelum ada peringkat
- Semi-Supervised Domain Adaptation Via Minimax Entropy PDFDokumen13 halamanSemi-Supervised Domain Adaptation Via Minimax Entropy PDFHưng NgôBelum ada peringkat
- Semi-Supervised Semantic Segmentation With Cross-Consistency TrainingDokumen18 halamanSemi-Supervised Semantic Segmentation With Cross-Consistency TrainingNamBelum ada peringkat
- Poster Yannan ShenDokumen1 halamanPoster Yannan ShenM Junaid SultanBelum ada peringkat
- Speech Compression Techniques: An OverviewDokumen4 halamanSpeech Compression Techniques: An OverviewEditor IJRITCCBelum ada peringkat
- OmniScan SW1.4 0603 Web PDFDokumen6 halamanOmniScan SW1.4 0603 Web PDFEur-Ing Nicola GalluzziBelum ada peringkat
- Monza 5 UHF Gen 2 RFID Tag Chip: IntroducingDokumen2 halamanMonza 5 UHF Gen 2 RFID Tag Chip: IntroducinggbbunevichBelum ada peringkat
- Improved Noise Discrimination For An Ammonia Tank AE Test: Specialist NDT ServicesDokumen28 halamanImproved Noise Discrimination For An Ammonia Tank AE Test: Specialist NDT ServicesGonzalo TelleríaBelum ada peringkat
- Distributed Arithmetic CodingDokumen10 halamanDistributed Arithmetic CodingperhackerBelum ada peringkat
- B5a 0605 00Dokumen25 halamanB5a 0605 00JS BBelum ada peringkat
- Bar Coding Tubular ProductsDokumen4 halamanBar Coding Tubular ProductsGusti PanjaitanBelum ada peringkat
- CM3-U3 Data SheetDokumen2 halamanCM3-U3 Data SheetmsmmcpbtergjmalueuBelum ada peringkat
- Multifocal Tutorial ZemaxDokumen8 halamanMultifocal Tutorial ZemaxDu RoyBelum ada peringkat
- Fiberpal OtdrDokumen2 halamanFiberpal OtdrAbdul AleemBelum ada peringkat
- NiceDokumen15 halamanNiceapi-3715782Belum ada peringkat
- Specim IQ Datasheet Short 02 WebDokumen2 halamanSpecim IQ Datasheet Short 02 WebJorge Hernán Osorio SánchezBelum ada peringkat
- Copernicus ProspektV5.0-UASDokumen2 halamanCopernicus ProspektV5.0-UASM.r SadrshiraziBelum ada peringkat
- Introduction To Computer Architecture and Network Tutorial 4.1 - Computer Component - Part 1: HardwareDokumen3 halamanIntroduction To Computer Architecture and Network Tutorial 4.1 - Computer Component - Part 1: HardwareTumblr AshwinBelum ada peringkat
- 1.2. LTE Cell PlanningDokumen35 halaman1.2. LTE Cell PlanningangdwipBelum ada peringkat
- Dynone ManualDokumen9 halamanDynone ManualLuisBelum ada peringkat
- Maize Sampler 2: DocumentationDokumen13 halamanMaize Sampler 2: DocumentationAnonymous Y1Htth0GqBelum ada peringkat
- Flexible OFDM Signal Generation, Analysis and TroubleshootingDokumen70 halamanFlexible OFDM Signal Generation, Analysis and TroubleshootingEdwin DrxBelum ada peringkat
- User Manual For TreeMix v1.0. Joseph K. Pickrell, Jonathan K. PritchardDokumen8 halamanUser Manual For TreeMix v1.0. Joseph K. Pickrell, Jonathan K. Pritchardstephenc144Belum ada peringkat
- EXFO Spec-Sheet AXS-110 v3 enDokumen3 halamanEXFO Spec-Sheet AXS-110 v3 enHuy DuBelum ada peringkat
- Vol7no3 331-336 PDFDokumen6 halamanVol7no3 331-336 PDFNghị Hoàng MaiBelum ada peringkat
- IrcamLab TS ManualDokumen30 halamanIrcamLab TS ManualShahaf HaverBelum ada peringkat
- Optimized Decoders For Mixed-Order AmbisonicsDokumen9 halamanOptimized Decoders For Mixed-Order AmbisonicsEric BenjaminBelum ada peringkat
- Unit-3 Lect MT Tele-2023Dokumen97 halamanUnit-3 Lect MT Tele-2023asfdSFDSFGEBelum ada peringkat
- Ftbx-735C Metro/Pon Fttx/Mdu OtdrDokumen7 halamanFtbx-735C Metro/Pon Fttx/Mdu Otdrsrc jatBelum ada peringkat
- 19EC406 Digital CommunicationDokumen4 halaman19EC406 Digital CommunicationGurunathanBelum ada peringkat
- Speaker Recognition Using Mel Frequency Cepstral Coefficients (MFCC) and VectorDokumen4 halamanSpeaker Recognition Using Mel Frequency Cepstral Coefficients (MFCC) and VectorAkah Precious ChiemenaBelum ada peringkat
- The Customized Network Pre-Planning Design Specification For Movitel - V0.0Dokumen17 halamanThe Customized Network Pre-Planning Design Specification For Movitel - V0.0Yordankys LópezBelum ada peringkat
- Jaideep jatinPPTDokumen12 halamanJaideep jatinPPTharshdeepbaby4Belum ada peringkat
- Convention Express Paper 36: Audio Engineering SocietyDokumen7 halamanConvention Express Paper 36: Audio Engineering SocietyŁukasz MorozBelum ada peringkat
- Lecture 4 - WDM NetworksDokumen15 halamanLecture 4 - WDM NetworksQadir MohtashamBelum ada peringkat
- Keonn-Advansafe-Data SheetDokumen3 halamanKeonn-Advansafe-Data SheetIrpal GiraseBelum ada peringkat
- Navigational Aids Audio CharacteristicsDokumen12 halamanNavigational Aids Audio Characteristicsspunk2meBelum ada peringkat
- Automatic Speaker Recognition For Mobile Communications Using Amr-Wb Speech CodingDokumen5 halamanAutomatic Speaker Recognition For Mobile Communications Using Amr-Wb Speech CodingBouhafs AbdelkaderBelum ada peringkat
- Microwave Journal-March 2021Dokumen168 halamanMicrowave Journal-March 2021Petros TsenesBelum ada peringkat
- Bio Drop-U LITEDokumen2 halamanBio Drop-U LITEParis100% (1)
- Evaluating Spatial Sound SystemsDokumen32 halamanEvaluating Spatial Sound SystemsMark BockoBelum ada peringkat
- DS ImagersCMOSDokumen2 halamanDS ImagersCMOSramadurruBelum ada peringkat
- SSP 5 3 Music CodingDokumen48 halamanSSP 5 3 Music CodingGabor GerebBelum ada peringkat
- Brosjyre Niton XL5Dokumen2 halamanBrosjyre Niton XL5Agustin A.Belum ada peringkat
- Ar New Technology With Matrix 320 enDokumen2 halamanAr New Technology With Matrix 320 enkuponator123Belum ada peringkat
- Speaker RecognitionDokumen15 halamanSpeaker RecognitionVignesh Vivekanandhan100% (1)
- Speech Recognition Over Digital Channels: Robustness and StandardsDari EverandSpeech Recognition Over Digital Channels: Robustness and StandardsBelum ada peringkat
- MPEG-7 Audio and Beyond: Audio Content Indexing and RetrievalDari EverandMPEG-7 Audio and Beyond: Audio Content Indexing and RetrievalBelum ada peringkat
- Kimball Et Al 2019, Figure 6Dokumen1 halamanKimball Et Al 2019, Figure 6Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 8Dokumen1 halamanKimball Et Al 2019, Figure 8Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 5Dokumen1 halamanKimball Et Al 2019, Figure 5Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 4Dokumen1 halamanKimball Et Al 2019, Figure 4Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 7Dokumen1 halamanKimball Et Al 2019, Figure 7Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 1Dokumen1 halamanKimball Et Al 2019, Figure 1Abby KimballBelum ada peringkat
- Tobin Et Al 2019, Figure 1Dokumen1 halamanTobin Et Al 2019, Figure 1Abby KimballBelum ada peringkat
- Tobin Et Al, 2019 Figure 3Dokumen1 halamanTobin Et Al, 2019 Figure 3Abby KimballBelum ada peringkat
- Neuwelt Et Al, Figure 2Dokumen1 halamanNeuwelt Et Al, Figure 2Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 4Dokumen1 halamanKimball Et Al 2019, Figure 4Abby KimballBelum ada peringkat
- Tobin Et Al 2019, Figure 2Dokumen1 halamanTobin Et Al 2019, Figure 2Abby KimballBelum ada peringkat
- Oko Et Al 2019, Figure 8Dokumen1 halamanOko Et Al 2019, Figure 8Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 1Dokumen1 halamanKimball Et Al 2019, Figure 1Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 10Dokumen1 halamanKimball Et Al 2018, Figure 10Abby KimballBelum ada peringkat
- Bullock Et Al 2018, Supplemental Figure 5Dokumen1 halamanBullock Et Al 2018, Supplemental Figure 5Abby KimballBelum ada peringkat
- Kimball Et Al 2019, Figure 3Dokumen1 halamanKimball Et Al 2019, Figure 3Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 9Dokumen1 halamanKimball Et Al 2018, Figure 9Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 3Dokumen1 halamanKimball Et Al 2018, Figure 3Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 2Dokumen1 halamanKimball Et Al 2018, Figure 2Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 8Dokumen1 halamanKimball Et Al 2018, Figure 8Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 7Dokumen1 halamanKimball Et Al 2018, Figure 7Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 5Dokumen1 halamanKimball Et Al 2018, Figure 5Abby KimballBelum ada peringkat
- Graph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectDokumen1 halamanGraph Features:: Events Sampled Per File: 9,141 Minimum Cluster Size: 5% Group Assignment: CorrectAbby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 4Dokumen1 halamanKimball Et Al 2018, Figure 4Abby KimballBelum ada peringkat
- Bullock Et Al 2018, Figure 6Dokumen1 halamanBullock Et Al 2018, Figure 6Abby KimballBelum ada peringkat
- Berger Et Al 2019, Figure 2Dokumen1 halamanBerger Et Al 2019, Figure 2Abby KimballBelum ada peringkat
- Kimball Et Al 2018, Figure 1Dokumen1 halamanKimball Et Al 2018, Figure 1Abby KimballBelum ada peringkat
- Recycling Mind MapDokumen2 halamanRecycling Mind Mapmsole124100% (1)
- All About PlantsDokumen14 halamanAll About Plantsapi-234860390Belum ada peringkat
- Furuno CA 400Dokumen345 halamanFuruno CA 400Димон100% (3)
- ChipmunkDokumen19 halamanChipmunkema.nemec13Belum ada peringkat
- Crashing Is A Schedule Compression Technique Used To Reduce or Shorten The Project ScheduleDokumen1 halamanCrashing Is A Schedule Compression Technique Used To Reduce or Shorten The Project ScheduleRaymart BulagsacBelum ada peringkat
- Mini Project 1 - 1Dokumen9 halamanMini Project 1 - 1Sameer BaraBelum ada peringkat
- Accuity: High Speed Automated 2D & 3D Optical Gauging Single Click Setup and InspectionDokumen4 halamanAccuity: High Speed Automated 2D & 3D Optical Gauging Single Click Setup and InspectionAPPLEBelum ada peringkat
- Class 12 - Maths - MatricesDokumen87 halamanClass 12 - Maths - MatricesAishwarya MishraBelum ada peringkat
- Rido, Rudini - Paediatric ECGDokumen51 halamanRido, Rudini - Paediatric ECGFikriYTBelum ada peringkat
- Modern Myth and Magical Face Shifting Technology in Girish Karnad Hayavadana and NagamandalaDokumen2 halamanModern Myth and Magical Face Shifting Technology in Girish Karnad Hayavadana and NagamandalaKumar KumarBelum ada peringkat
- S590 Machine SpecsDokumen6 halamanS590 Machine SpecsdilanBelum ada peringkat
- Ge Fairchild Brochure PDFDokumen2 halamanGe Fairchild Brochure PDFDharmesh patelBelum ada peringkat
- Vehicle Intercom Systems (VIS)Dokumen4 halamanVehicle Intercom Systems (VIS)bbeisslerBelum ada peringkat
- Top Coat-200 - Data PDFDokumen4 halamanTop Coat-200 - Data PDFLiliana GeorgianaBelum ada peringkat
- Exercise 9 Two Factor Factorial Experiments and Derivation of Expected Mean SquaresDokumen14 halamanExercise 9 Two Factor Factorial Experiments and Derivation of Expected Mean SquaresHasmaye PintoBelum ada peringkat
- ReviewerDokumen3 halamanReviewerKristine SantominBelum ada peringkat
- Igcse Revision BookDokumen23 halamanIgcse Revision BookJo Patrick100% (2)
- Chapter 10 - The Mature ErythrocyteDokumen55 halamanChapter 10 - The Mature ErythrocyteSultan AlexandruBelum ada peringkat
- 1.1 The Prescription of Blood ComponentsDokumen9 halaman1.1 The Prescription of Blood ComponentsagurtovicBelum ada peringkat
- Sw34 Religion, Secularism and The Environment by NasrDokumen19 halamanSw34 Religion, Secularism and The Environment by Nasrbawah61455Belum ada peringkat
- Upaam 1135891 202105060749199700Dokumen18 halamanUpaam 1135891 202105060749199700Kartik KapoorBelum ada peringkat
- ZhentarimDokumen4 halamanZhentarimLeonartBelum ada peringkat
- Aesculap Saw GD307 - Service ManualDokumen16 halamanAesculap Saw GD307 - Service ManualFredi PançiBelum ada peringkat
- Comparison of 3 Tests To Detect Acaricide ResistanDokumen4 halamanComparison of 3 Tests To Detect Acaricide ResistanMarvelous SungiraiBelum ada peringkat
- Product Stock Exchange Learn BookDokumen1 halamanProduct Stock Exchange Learn BookSujit MauryaBelum ada peringkat
- Chapter Three: 1 - The Coarse Grain SoilsDokumen21 halamanChapter Three: 1 - The Coarse Grain SoilsSalih MohayaddinBelum ada peringkat
- Ventricular Septal DefectDokumen9 halamanVentricular Septal DefectpepotchBelum ada peringkat
- 9701 w09 QP 21Dokumen12 halaman9701 w09 QP 21Hubbak KhanBelum ada peringkat
- Moses ManualDokumen455 halamanMoses ManualDadypeesBelum ada peringkat
- Abdominal Examination OSCE GuideDokumen30 halamanAbdominal Examination OSCE Guideزياد سعيدBelum ada peringkat