Antibiotic Resistance: Microbiology: A Clinical Approach © Garland Science
Diunggah oleh
Anonymous Ra09vhgLJudul Asli
Hak Cipta
Format Tersedia
Bagikan dokumen Ini
Apakah menurut Anda dokumen ini bermanfaat?
Apakah konten ini tidak pantas?
Laporkan Dokumen IniHak Cipta:
Format Tersedia
Antibiotic Resistance: Microbiology: A Clinical Approach © Garland Science
Diunggah oleh
Anonymous Ra09vhgLHak Cipta:
Format Tersedia
Microbiology: A Clinical Approach Garland Science
ANTIBIOTIC RESISTANCE
WHY IS THIS IMPORTANT?
If our antibiotics stop working what will happen
to people when they get sick?
It will force us to change the way we view
disease and the way we treat patients.
ANTIBIOTIC RESISTANCE
Antibiotics use has not been without consequence.
There are several factors in the development of
antibiotic resistance:
Overuse Doctors first line is to give antibiotics and not
let body do its job!
Misuse Doctors prescribe antibiotic for a
virus.antibiotics only kill bacteria
Not finishing people dont finish medicine
Technology people travel more and spread more
HOW IS IT HAPPENING?
Bacterial cells that have developed resistance are
not killed by medicine.
They continue to divide and pass on their resistance to
their offspring.
The new group of bacteria cant be killed by that drug.
Penicillin G
Amoxicillin Ampicillin Azlocillin Carbenicillin Cloxacillin Dicloxacillin Flucloxacillin Mezlocillin
Methicillin Nafcillin
Oxacillin Piperacillin Temocillin Ticarcillin
Amoxicillin/clavulanate Ampicillin/sulbactam
Piperacillin/tazobactam Ticarcillin/clavulanate
EVOLUTION OF RESISTANCE TO
ANTIBIOTIC
..DEVELOPMENT OF RESISTANCE
TODAY
1. Find petri dishes and
make a note of the
colonies (# and size)
2. Get a small beaker,
add a small white paper
disc + 1 dropper full of
cleaning solution
3. Soak for 5 minutes
4. Use tweezers to put
that disc near some of
the growing colonies.
TECHNOLOGY, BACTERIAS BFF
Modern technology and sociology is also
increasing antibiotic resistance
Travelers may carry resistant bacteria from one
country to another
They infect Other people with the resistant
bacteria.
The process is repeated and the resistant bacteria
spread.
LIVING CONDITIONS
There are more large cities in the world today.
Large numbers of people in relatively small
areas
Passing antibiotic-resistant pathogens is easier.
Many large urban populations have poor
sanitation.
DEVELOPMENT OF RESISTANCE:
FOOD
Food is also a source of infection that could
affect the development of resistance.
More meals are prepared outside the home.
Contamination goes unnoticed until infection has
started.
Outbreaks of Escherichia coli O157 in spinach and
lettuce in the US.
As the number of foodborne infections increases,
so does the use of antibiotics.
Causes an increase in the development of resistance.
EVOLUTION OF RESISTANCE TO
ANTIBIOTIC
..EVOLUTION OF ANTIBIOTIC
RESISTANCE: RATE OF DEVELOPMENT
Resistance develops at different rates.
Several groups of antibiotics were used
for many years before resistance was
seen.
Resistance to penicillin was seen in
only three years.
..EVOLUTION OF ANTIBIOTIC
RESISTANCE: RATE OF DEVELOPMENT
..EVOLUTION OF ANTIBIOTIC
RESISTANCE: OVERUSE
The most important contributing factor for
resistance is overuse.
A good example is prescribing antibiotics that
dont kill viruses for the common cold.
These antibiotics do destroy the normal flora.
Opportunistic pathogens that are resistant
survive and can take hold.
EVOLUTION OF ANTIBIOTIC RESISTANCE:
HOSPITALS
Hospitals are ideal reservoirs for the acquisition of
resistance.
A population of people with compromised health
A high concentration of organisms, many of which are
extremely pathogenic
Large amounts of different antibiotics are constantly in
use
Increased use of antibiotics leads to resistance.
Hospital is a place where resistance can develop rapidly.
..EVOLUTION OF ANTIBIOTIC
RESISTANCE: TRANSFER
Resistance can be transferred by bacteria
swapping genes.
This can be easily accomplished in a hospital setting.
Health care workers who dont follow infection
control protocols aid in increasing resistance.
..EVOLUTION OF ANTIBIOTIC
RESISTANCE: RESISTANCE ISLANDS
Plasmids containing genes for resistance can
integrate into the chromosome.
Here they form resistance islands.
Resistance genes accumulate and are stably
maintained.
..EVOLUTION OF ANTIBIOTIC
RESISTANCE
Microorganisms producing antibiotic substances
have autoprotective mechanisms.
Transmembrane proteins pump out the freshly
produced antibiotic so that it does not accumulate.
If it did, it would kill the organism producing it.
Genes that code for these pumps are closely
linked to genes that code for antibiotic
substances.
When genes for antibiotic production are turned on so
are the pump genes.
MECHANISMS FOR ACQUIRING
RESISTANCE
Bacteria use several mechanisms to become
antibiotic-resistant:
Inactivation of the antibiotic
Efflux pumping of the antibiotic
Modification of the antibiotic target
Alteration of the pathway
INACTIVATION OF ANTIBIOTIC
Inactivation involves enzymatic breakdown of
antibiotic molecules.
A good example is -lactamase:
Secreted into the bacterial periplasmic space
Attacks the antibiotic as it approaches its target
There are more than 190 forms of -lactamase.
E.g of lactamase activity in E.coli and S. aureus (read
pg 469).
MECHANISMS FOR ACQUIRING
RESISTANCE
EFFLUX PUMPING OF ANTIBIOTIC
Efflux pumping is an active transport mechanism.
It requires ATP.
Efflux pumps are found in:
The bacterial plasma membrane
The outer layer of gram-negative organisms
Pumping keeps the concentration of antibiotic below
levels that would destroy the cell
Genes that code for efflux pumps are located on plasmids
and transposons.
Transposons are sequences of DNA that can move or
transpose move themselves to new positions within the
genome of a single cell.Transposones:
Readily acquired by nonresistant bacteria
Transforms them into resistant bacteria
OTHER MECHANISMS OF KEEPING
ANTIBIOTICS OUT
Some bacteria reduce the permeability of their
membranes as a way of keeping antibiotics out.
They turn off production of porin and other membrane
channel proteins.
Seen in resistance to streptomycin, tetracycline, and
sulfa drugs.
MODIFICATION OF ANTIBIOTIC TARGET
Bacteria can modify the antibiotics target to
escape its activity (pg470)
Bacteria must change structure of the target but
the modified target must still be able to function.
This can be achieved in two ways:
Mutation of the gene coding for the target protein
Importing a gene that codes for a modified target
E.g. with MRSA (methicillin- resistant - S. aureus ),
similar to PBP (penicillin- binding- protein)
..MODIFICATION OF ANTIBIOTIC TARGET
Bacteria have PBPs in their plasma membranes.
These proteins are targets for penicillin.
MRSA has acquired a gene (mec A) that codes for
a different PBP.
It has a different three-dimensional structure.
MRSA less sensitive to penicillins.
..MODIFICATION OF ANTIBIOTIC TARGET
MRSA is resistant to all -lactam antibiotics,
cephalosporins, and carbapenems.
It is a very dangerous pathogen particularly in burn
patients
Streptococcus pneumoniae also modifies PBP.
It can make as many as five different types of PBP.
It does this by rearranging, or shuffling, the genes.
Referred to as genetic plasticity
Permits increased resistance
MODIFICATION OF TARGET RIBOSOMES
Bacterial ribosomes are a primary target for
antibiotics (pg471).
Different antibiotics affect them in different ways.
Resistance can be the result of modification of
ribosomal RNA so it is no longer sensitive.
Some organisms use target modification in
conjunction with efflux pumps.
Resistance is even more effective.
ALTERATION OF A PATHWAY
Some drugs competitively inhibit metabolic
pathways.
Bacteria can overcome this method by using an
alternative pathway.
Approximately 7% of the total S. aureus genome
is genes for antibiotic resistance.
Bacillus subtilis, a nonpathogenic organism, has
none.
..ALTERATION OF A PATHWAY
Several specific resistance genes of MRSA have
been identified. They are associated with different
resistance mechanisms.
-lactamase resistance
Erythromycin resistance
Production of aminoglycosides
Operation of efflux pumps
..ALTERATION OF A PATHWAY
Bacteria that are part of the normal flora are
becoming more dangerous due to resistance.
E. coli is part of the normal flora of the large intestine.
It has become more involved with urinary tract
infections.
Antibiotic-resistant infections are now being seen
throughout the world.
MRSA, VRSA, VRE, AND OTHER
PATHOGENS
Several antibiotic-resistant bacteria are
considered clinically dangerous.
MRSA (Methicillin-Resistant ) and VRSA
(Vancomycin-resitant S.aureus are very virulent in
humans and are referred as professional pathogens.
(Refer Table 20.3 pg 472)
MRSA and VRSA contain many resistance
genes.
Three or four resistance islands on the chromosome
26-28 additional gene clusters on plasmids which can
move to other bacterial cells.
VRE-Vancomycin enterococcus e.g E. faecalis
contributes to 90% of all vancomycin resistant
bacteria
CONTRIBUTING FACTORS AND POSSIBLE
SOLUTIONS
Variety of factors involved in the development of
drug resistance (Pg 474)
An underestimated factor is the success of so
many antibiotics.
They are very effective at killing microorganisms.
Doctors and patients have become dependent on them.
CONTRIBUTING FACTORS AND POSSIBLE
SOLUTIONS
The doctor-patient-drug relationship leads to
resistance.
Most clearly seen in the case of common viral
infections.
Patients expect to have antibiotics have prescribed.
There is overprescription of antibiotics that are not
required.
Patients who feel better and stop using the drug make
the problem worse.
CONTRIBUTING FACTORS AND POSSIBLE
SOLUTIONS
Overuse of broad-spectrum antibiotics
(cephalosporins) leads to the rise of resistance.
It permits the superinfection effect.
Pathogens occupy areas where normal microbes have been
killed.
Antibiotics have essentially compromised the patient.
CONTRIBUTING FACTORS AND POSSIBLE
SOLUTIONS
Clostridium difficile is a superinfection pathogen.
Establishes itself in the intestinal tract as part of a
superinfection
It is very resistant to antibiotics.
Patients with this infection are difficult to treat
DESTRUCTION OF NORMAL FLORA
ALLOWS PATHOGENIC PATHOGENS TO
DOMINATE
..CONTRIBUTING FACTORS AND
POSSIBLE SOLUTIONS
The potential for global antibiotic resistance is
real due to:
Overuse of antibiotics
Improper adherence to hospital infection control
protocols
Difficulty finding new antibiotics
Ease of worldwide travel
There are ways to lengthen the useful life of
antibiotics.
GUIDELINES FOR EXTENDING THE
USEFUL LIFE OF ANTIMICROBIAL DRUGS
REFERENCES
Microbiology, A clinical Approach -
Danielle Moszyk-Strelkauskas-Garland
Science 2010
http://en.wikipedia.org/wiki/Scientific_met
hod
Anda mungkin juga menyukai
- History: The Problem: Genetic Evolution?Dokumen14 halamanHistory: The Problem: Genetic Evolution?Sumeena VasundhraBelum ada peringkat
- Sajb 55334 336Dokumen3 halamanSajb 55334 336Joe HaBelum ada peringkat
- AntibioticsDokumen5 halamanAntibioticsSneeha VeerakumarBelum ada peringkat
- Antibiotic Resistant Bacteria ThesisDokumen4 halamanAntibiotic Resistant Bacteria Thesislauramillerscottsdale100% (2)
- Hello Bio InvestDokumen16 halamanHello Bio InvestAadrica WaliaBelum ada peringkat
- Antibiotic ResistanceDokumen29 halamanAntibiotic ResistanceDian Aditama100% (1)
- IntroductionDokumen12 halamanIntroductionSuvidVijay FadanavisBelum ada peringkat
- Antibiotics & Antibiotic ResistanceDokumen53 halamanAntibiotics & Antibiotic ResistanceLeenoos RayapanBelum ada peringkat
- Biology Project XiiDokumen14 halamanBiology Project XiiSagayaraniBelum ada peringkat
- Resistencia Antimicrobiana en InglesDokumen14 halamanResistencia Antimicrobiana en Inglesstefanymoreno22Belum ada peringkat
- Mechanisms of Antimicrobial Resistance in Bacteria PDFDokumen8 halamanMechanisms of Antimicrobial Resistance in Bacteria PDFJuan Pablo Ribón GómezBelum ada peringkat
- Name: Kiran Bhatti University: Liaquat University of Medical and Health Sciences. Pharm-D StudentDokumen9 halamanName: Kiran Bhatti University: Liaquat University of Medical and Health Sciences. Pharm-D StudentPharmanic By Ruttaba FatimaBelum ada peringkat
- Bio ProjectDokumen6 halamanBio ProjectAKM KINGBelum ada peringkat
- BIOLOGY Practical File For Class 12Dokumen12 halamanBIOLOGY Practical File For Class 12onlyaimiitBelum ada peringkat
- Trends in Antibiotic Therapy: Rt. Rev. Prof. C.S.S. BelloDokumen47 halamanTrends in Antibiotic Therapy: Rt. Rev. Prof. C.S.S. BelloanneBelum ada peringkat
- Antibiotics Drug ResistanceDokumen10 halamanAntibiotics Drug ResistancePrithvi de VilliersBelum ada peringkat
- Antibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentDokumen35 halamanAntibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentRoby Aditya SuryaBelum ada peringkat
- Antimicrobial Resistances: The World's Next Pandemic On The WayDokumen11 halamanAntimicrobial Resistances: The World's Next Pandemic On The WayIAEME PublicationBelum ada peringkat
- Antimicrobial TherapyDokumen18 halamanAntimicrobial TherapysaifBelum ada peringkat
- Noakhali Science & Technology University: Assignment OnDokumen18 halamanNoakhali Science & Technology University: Assignment OnSubrina ChowdhuryBelum ada peringkat
- Antibiotic Resistance Thesis StatementDokumen6 halamanAntibiotic Resistance Thesis Statementkatelogebellevue100% (2)
- Chapter 9 Surgical Infections and Antibiotic SelectionDokumen42 halamanChapter 9 Surgical Infections and Antibiotic SelectionSteven Mark MananguBelum ada peringkat
- Presentation On Antibiotic: By:-Shiv Kumar Roll No. 21 Mba BTDokumen18 halamanPresentation On Antibiotic: By:-Shiv Kumar Roll No. 21 Mba BTAnshuman ParasharBelum ada peringkat
- Ijpab 2014 2 3 207 226 PDFDokumen20 halamanIjpab 2014 2 3 207 226 PDFAlmuja dilla Ulta DeputriBelum ada peringkat
- SuperbugsDokumen3 halamanSuperbugsMatei BuneaBelum ada peringkat
- Jurnal AntibiotikDokumen16 halamanJurnal AntibiotikMaulida 04Belum ada peringkat
- Morrison 2020Dokumen17 halamanMorrison 2020ANGEL DANIEL CASTAÑEDA PAREDESBelum ada peringkat
- 2003 Antibiotic Selection in Head and Neck InfectionsDokumen22 halaman2003 Antibiotic Selection in Head and Neck InfectionsMariana ZebadúaBelum ada peringkat
- Abstract 1Dokumen6 halamanAbstract 1mv chinmayBelum ada peringkat
- Research Paper Antibiotic ResistanceDokumen5 halamanResearch Paper Antibiotic Resistancegvyns594100% (1)
- Bio Project Drug Resistance in BacteriaDokumen18 halamanBio Project Drug Resistance in BacteriaAKASH ALAMBelum ada peringkat
- Antibiotic Resistance and The New Antibiotic Agents: SV Want, A HolmesDokumen7 halamanAntibiotic Resistance and The New Antibiotic Agents: SV Want, A Holmesudaysingh98Belum ada peringkat
- Bio ProjectDokumen16 halamanBio ProjectmuddurajusrpBelum ada peringkat
- HNS 2202 Lesson 3Dokumen21 halamanHNS 2202 Lesson 3hangoverBelum ada peringkat
- ANTIMICROBIALS (Israjaved)Dokumen23 halamanANTIMICROBIALS (Israjaved)Isra JavedBelum ada peringkat
- Origins of Antibiotics: Aureus) Had Disappeared. He Realized That Not Only Did This Mold-WhichDokumen11 halamanOrigins of Antibiotics: Aureus) Had Disappeared. He Realized That Not Only Did This Mold-WhichAndreas StavrianosBelum ada peringkat
- Bio Project Calss 11Dokumen20 halamanBio Project Calss 11muddurajusrpBelum ada peringkat
- Resistência Antimicrobiana - MandellDokumen21 halamanResistência Antimicrobiana - MandellRafaelGomesdeMeloBelum ada peringkat
- Antibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentDokumen12 halamanAntibiotic Resistance and Extended Spectrum Beta-Lactamases: Types, Epidemiology and TreatmentyomnayasminBelum ada peringkat
- Multi Drug ResistanceDokumen33 halamanMulti Drug ResistanceEvanslin SantusBelum ada peringkat
- Super Bug - Unmaskingthethreat WootensalkindDokumen14 halamanSuper Bug - Unmaskingthethreat WootensalkindMatei BuneaBelum ada peringkat
- Assignment O1Dokumen17 halamanAssignment O1irfanh15951Belum ada peringkat
- Assignment On:: Antibiotic Resistance and Bacterial Biofilm: Causes and ThreatsDokumen7 halamanAssignment On:: Antibiotic Resistance and Bacterial Biofilm: Causes and ThreatsArafat MiahBelum ada peringkat
- Biology Project Drug Resistance To BacteriaDokumen6 halamanBiology Project Drug Resistance To BacteriaAyush YadavBelum ada peringkat
- Antibiotic Resistance Mechanisms in Bacteria: Biochemical and Genetic AspectsDokumen11 halamanAntibiotic Resistance Mechanisms in Bacteria: Biochemical and Genetic AspectsD Wisam NajmBelum ada peringkat
- Microbiology - Exercise 3e AntibiogramDokumen5 halamanMicrobiology - Exercise 3e Antibiogramapi-253346521Belum ada peringkat
- INDEXDokumen16 halamanINDEXayush rajeshBelum ada peringkat
- 1 s2.0 S097594761630448X MainDokumen10 halaman1 s2.0 S097594761630448X MainSamBelum ada peringkat
- Antimicrobial Susceptibility Testing (AST)Dokumen41 halamanAntimicrobial Susceptibility Testing (AST)summiya100% (1)
- Antibiotic Resistance: Dr. Sharifah Shakinah Supervisor: Dr. Eppy SPPD KptiDokumen25 halamanAntibiotic Resistance: Dr. Sharifah Shakinah Supervisor: Dr. Eppy SPPD KptiShasha Shakinah100% (1)
- Mechanisms of Bacterial Resistance: Dr. Hawa DC 500 For DDS 2017/2018Dokumen25 halamanMechanisms of Bacterial Resistance: Dr. Hawa DC 500 For DDS 2017/2018DavidBelum ada peringkat
- Antibiotic Resistance Genes: Research Project TitleDokumen14 halamanAntibiotic Resistance Genes: Research Project TitleHassaninElaradyBelum ada peringkat
- Drug Resistance Pharmacology Lecture Lecture Series 2019Dokumen23 halamanDrug Resistance Pharmacology Lecture Lecture Series 2019temitopeBelum ada peringkat
- The Study of Drug Resistance in Bacteria Using Antibiotics: June 2019Dokumen6 halamanThe Study of Drug Resistance in Bacteria Using Antibiotics: June 20197'A'06Aditya BeheraBelum ada peringkat
- 1 s2.0 S1198743X16000288 MainDokumen3 halaman1 s2.0 S1198743X16000288 MainAizaz HassanBelum ada peringkat
- Antibiotic Use: Present and Future: Stephen H. ZinnerDokumen5 halamanAntibiotic Use: Present and Future: Stephen H. Zinnermbilouzi589Belum ada peringkat
- Case Study of MicrobiologyDokumen4 halamanCase Study of MicrobiologyQaisrani Y9Belum ada peringkat
- Haider Antibiotic Resistance 2023-24Dokumen42 halamanHaider Antibiotic Resistance 2023-24R. nounBelum ada peringkat
- PHARMACOLOGY AntibioticsDokumen29 halamanPHARMACOLOGY AntibioticsBeck33ers5826100% (1)
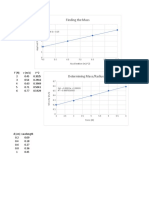
- Finding The Mass: F (N) A (M/s 2) 4.5 2 6.5 3 8.5 4 10.5 5Dokumen2 halamanFinding The Mass: F (N) A (M/s 2) 4.5 2 6.5 3 8.5 4 10.5 5Anonymous Ra09vhgLBelum ada peringkat
- 13 Frog Dissection Lab-1 Done ADokumen12 halaman13 Frog Dissection Lab-1 Done AAnonymous Ra09vhgLBelum ada peringkat
- LogDokumen39 halamanLogAnonymous Ra09vhgLBelum ada peringkat
- Genetics: Inheritance of AlbinismDokumen13 halamanGenetics: Inheritance of AlbinismAnonymous Ra09vhgL0% (1)
- Graphic Diagram Sexual ReproductionDokumen1 halamanGraphic Diagram Sexual ReproductionAnonymous Ra09vhgLBelum ada peringkat
- Centers For Disease Control and Prevention. Centers For Disease Control and Prevention, N.DDokumen1 halamanCenters For Disease Control and Prevention. Centers For Disease Control and Prevention, N.DAnonymous Ra09vhgLBelum ada peringkat
- Introduction To Thermodynamics: I. Conservation of EnergyDokumen9 halamanIntroduction To Thermodynamics: I. Conservation of EnergyAnonymous Ra09vhgLBelum ada peringkat
- What Are Stem Cells and Where Do They Come FromDokumen4 halamanWhat Are Stem Cells and Where Do They Come FromjaniellegbBelum ada peringkat
- Cell Venn Diagram Plants Vs AnimalsDokumen2 halamanCell Venn Diagram Plants Vs AnimalsAnonymous Ra09vhgLBelum ada peringkat
- Frog Anatomy 1Dokumen40 halamanFrog Anatomy 1Anonymous Ra09vhgLBelum ada peringkat
- Evolution of The HeartDokumen12 halamanEvolution of The HeartAnonymous Ra09vhgLBelum ada peringkat
- Páginas de Rockwood Fraturas em Adultos, 8ed3Dokumen51 halamanPáginas de Rockwood Fraturas em Adultos, 8ed3Estevão CananBelum ada peringkat
- What Is Malnutrition?: WastingDokumen6 halamanWhat Is Malnutrition?: WastingĐoan VõBelum ada peringkat
- Environmental Studies-FIRST UNIT-vkmDokumen59 halamanEnvironmental Studies-FIRST UNIT-vkmRandomBelum ada peringkat
- Acid Base Disorders JAPIDokumen5 halamanAcid Base Disorders JAPIVitrag_Shah_1067Belum ada peringkat
- Conduction Blocks in Acute Myocardial Infarction: A Prospective StudyDokumen6 halamanConduction Blocks in Acute Myocardial Infarction: A Prospective StudyJack JacksonBelum ada peringkat
- Test Bank For Microbiology Basic and Clinical Principles 1st Edition Lourdes P Norman MckayDokumen36 halamanTest Bank For Microbiology Basic and Clinical Principles 1st Edition Lourdes P Norman Mckayshriekacericg31u3100% (43)
- Tendonitis Guide - Causes, Symptoms and Treatment OptionsDokumen6 halamanTendonitis Guide - Causes, Symptoms and Treatment OptionsSylvia GraceBelum ada peringkat
- Histopathology of Dental CariesDokumen7 halamanHistopathology of Dental CariesJOHN HAROLD CABRADILLABelum ada peringkat
- Cage HomesDokumen8 halamanCage HomesstanhopekrisBelum ada peringkat
- Para Lab 11Dokumen3 halamanPara Lab 11api-3743217Belum ada peringkat
- Bipolar Disorder - A Cognitive Therapy Appr - Cory F. NewmanDokumen283 halamanBipolar Disorder - A Cognitive Therapy Appr - Cory F. NewmanAlex P100% (1)
- Malawi Clinical HIV Guidelines 2019 Addendumversion 8.1Dokumen28 halamanMalawi Clinical HIV Guidelines 2019 Addendumversion 8.1INNOCENT KHULIWABelum ada peringkat
- Parathyroi D HormoneDokumen25 halamanParathyroi D HormoneChatie PipitBelum ada peringkat
- Peoples of Cambodia 2nd EditionDokumen55 halamanPeoples of Cambodia 2nd Editionsummit_go_team100% (1)
- Beatriz Colomina X Ray Architecture Lars Muller (046 090)Dokumen45 halamanBeatriz Colomina X Ray Architecture Lars Muller (046 090)SABUESO FINANCIEROBelum ada peringkat
- Algorithm: Essential Newborn Care: Is The Baby Gasping or Not Breathing?Dokumen1 halamanAlgorithm: Essential Newborn Care: Is The Baby Gasping or Not Breathing?Dela cruz KimberlyBelum ada peringkat
- DSM OcdDokumen2 halamanDSM Ocdnmyza89Belum ada peringkat
- Subjective: Diarrhea Related To Watery Short Term: IndependentDokumen4 halamanSubjective: Diarrhea Related To Watery Short Term: IndependentEmma Lyn SantosBelum ada peringkat
- Biology Combined Science Notes: Changamire Farirayi D.T. Form 3 & 4 BiologyDokumen67 halamanBiology Combined Science Notes: Changamire Farirayi D.T. Form 3 & 4 BiologyKeiron Cyrus100% (6)
- 1.11 ANATOMY - The Nose and Paransal SinusesDokumen4 halaman1.11 ANATOMY - The Nose and Paransal SinusesPaolo NaguitBelum ada peringkat
- GMO Pro Arguments (Gen-WPS OfficeDokumen4 halamanGMO Pro Arguments (Gen-WPS OfficeFranchesca RevelloBelum ada peringkat
- Illness in The NewbornDokumen34 halamanIllness in The NewbornVivian Jean TapayaBelum ada peringkat
- Development of The Vertebrate EyeDokumen5 halamanDevelopment of The Vertebrate Eyezari_pak2010Belum ada peringkat
- 2 - Cannabis Intoxication Case SeriesDokumen8 halaman2 - Cannabis Intoxication Case SeriesPhelipe PessanhaBelum ada peringkat
- Journals With No Article Processing ChargesDokumen14 halamanJournals With No Article Processing ChargesNurul Jami Bakti JayaBelum ada peringkat
- Aluminum in Co Ffee: AccessDokumen9 halamanAluminum in Co Ffee: AccessJohnBelum ada peringkat
- MBF90324234Dokumen407 halamanMBF90324234Emad Mergan100% (1)
- Reviewer in MCNDokumen16 halamanReviewer in MCNChristian Clyde Noel JakosalemBelum ada peringkat
- Sample ReportDokumen3 halamanSample ReportRobeants Charles PierreBelum ada peringkat
- Canadian Pharmacy Review Ver1Dokumen6 halamanCanadian Pharmacy Review Ver1Dr-Usman Khan30% (10)